Resonance Assignment/AutoAssign
Introduction
AutoAssign is a constraint-based expert system for automating the assignment and analysis of backbone NMR resonance assignments of proteins (Ref. 1,2). The program is implemented in C++, Java2, and Perl programming languages and supported on SGI-IRIX, Sun-Solaris, MAC-OSX, x86-Linux, and x86_64-Linux architectures. The newest AutoAssign distribution (version 2.4.0) automates the assignments of HN, NH, CO, CA, CB, HA, and HB resonances in non-, partially-, and fully-deuterated samples. The rich graphical user interface (GUI) provides a many sets of tools for dataset conversions, assignment validations, and various graphical displays of assignment results. AutoAssign is well tested on a large number of independently-collected triple-resonance NMR data sets of proteins ranging in size from ~6 to ~32 kD, including one fully-deuterated protein and and a dataset with reduced-dimensionality experiments. AutoAssign performs the automated analysis of assignments in only seconds on current RISC and x86 platforms.
The following description is mainly taken from the AutoAssign SOP (tutorial). Please check this tutorial and the version 2.4.0 help documentation for more information and usage.
Using AutoAssign
Input Files
The input files for AutoAssign are:
- A sequence file in fasta format
- Peak lists for backbone experiments in Sparky format.
Opening AutoAssign
- Run the startup script
autoassignIf you are running the client on the same machine as the server. - If you are running the client on a machine different from the server, then you must first make sure that the server is running on the other machine. The easiest way to do this is to run the startup script
autoserveron this other computer.
The main AutoAssign window will appear (Figure 1).
Figure 1: AutoAssign Main Window
Creating a Data Set
Using the Peaklist Converter in the Graphical User Interface
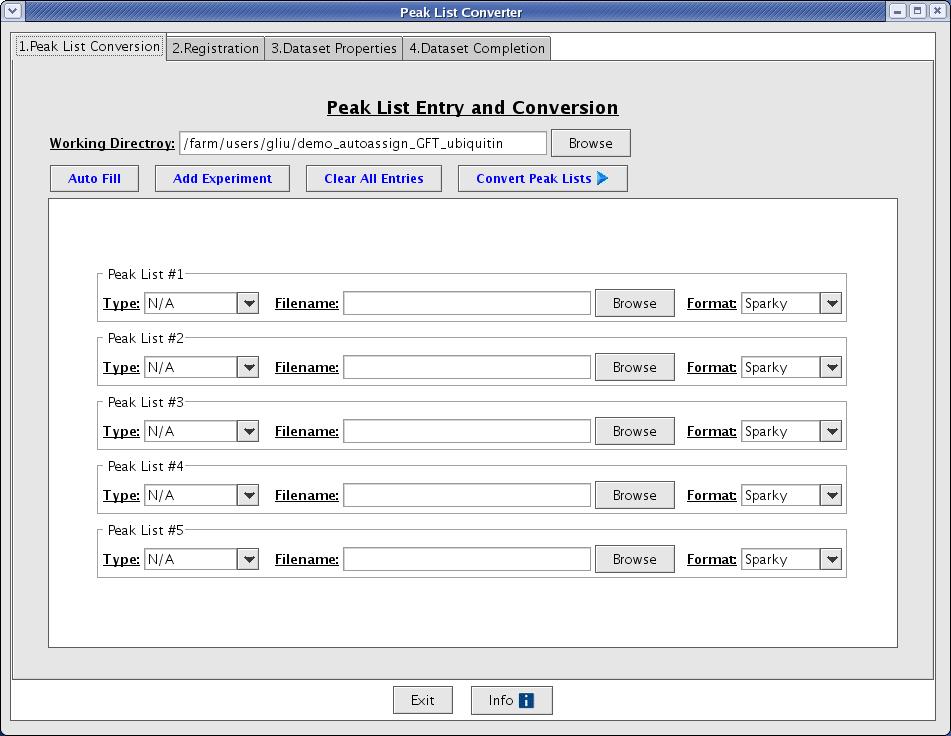
The easiest way to create a dataset is to using the Dataset Creation Graphical User Interface. This interface will guide you through the basic steps of converting your peak lists, registering your peak lists, creating a control file, and evaluating the quality of your peak lists. Go to File -> Convert/Create Dataset from the main menu option to start the Dataset Creation Graphical User Interface (Figure 2).
Figure 2: The Peak List Converter
The process is divided into 4 steps: 1. Peak list conversion, 2. Registration, 3. Data set properties, and 4. Data set completion.
Peak list conversion
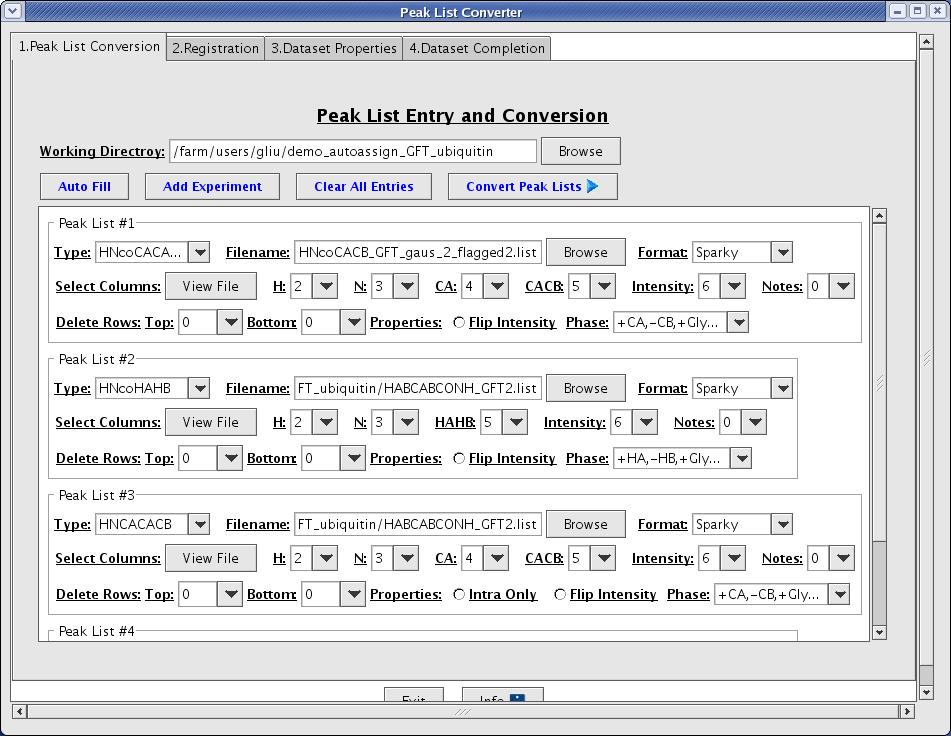
The peak list conversion process is designed to reformat given peak lists into AutoAssign format (Figure 3). Here the user defines the columns and spectral properties of each peak list.
Figure 3: Peak List Conversion Page
- Working directory [text field]: it is a base directory that the interface will use to create a subdirectory with a time stamp. The default is set to the current working directory.
- Auto Fill [button]: when a user decides to reuse the same peak lists or the same type of configuration based on the previous analysis, this function allows a user to import the data previously used.
- Add Experiment [button]: This function allows a user to display additional blank peaklist panel.
- Clear All Entries [button]: this function clears populated data fields displayed on screens.
- Convert Peak Lists [button]: this function runs a set of AutoAssign perl scripts to converts peak lists entered by a user. It generates intermediate files used to compute registration/tolerance values for the subsequent steps. All scripts executions run by interface is logged in
event.logfile found under the subdirectory named with a time stamp along with other intermediate files.
Registration
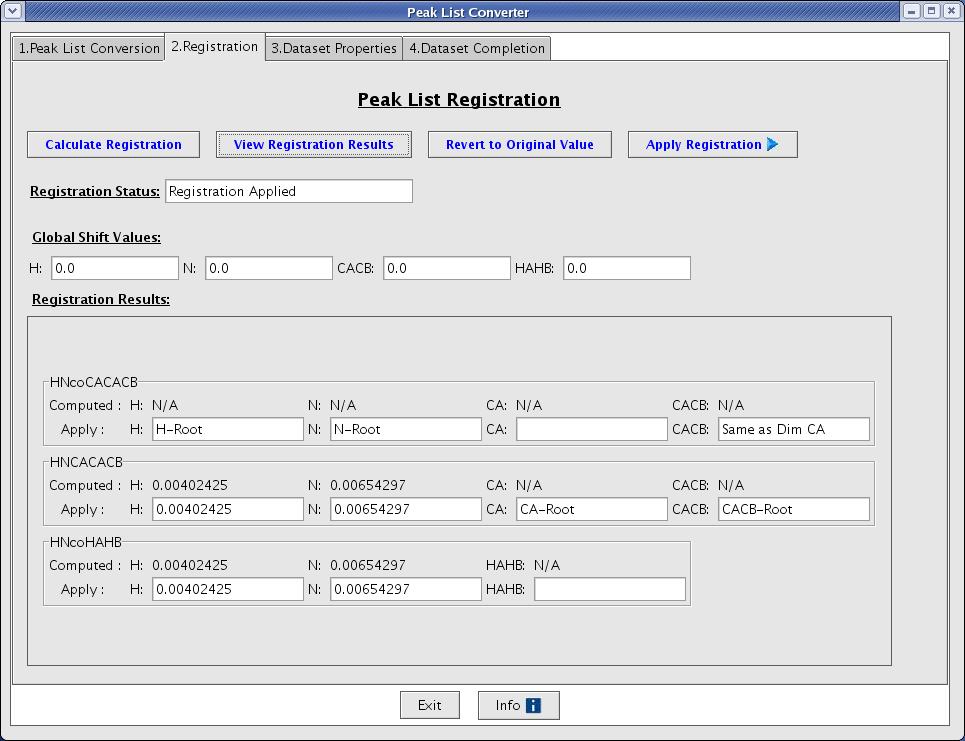
A user will be taken to the Registration tab when Convert Peak List button is clicked. This will allow a user to calculate registration values and apply those values to shift peaks by creating new / temporal peaklist files (Figure 4).
Figure 4: Peak List Registration Page
- Calculate Registration [button]: this function starts calculating registration value and it will open a progress monitor window to notify you how many calculations are completed. The error may be recorded in .reg file(s) under subdirectory if it occurs.
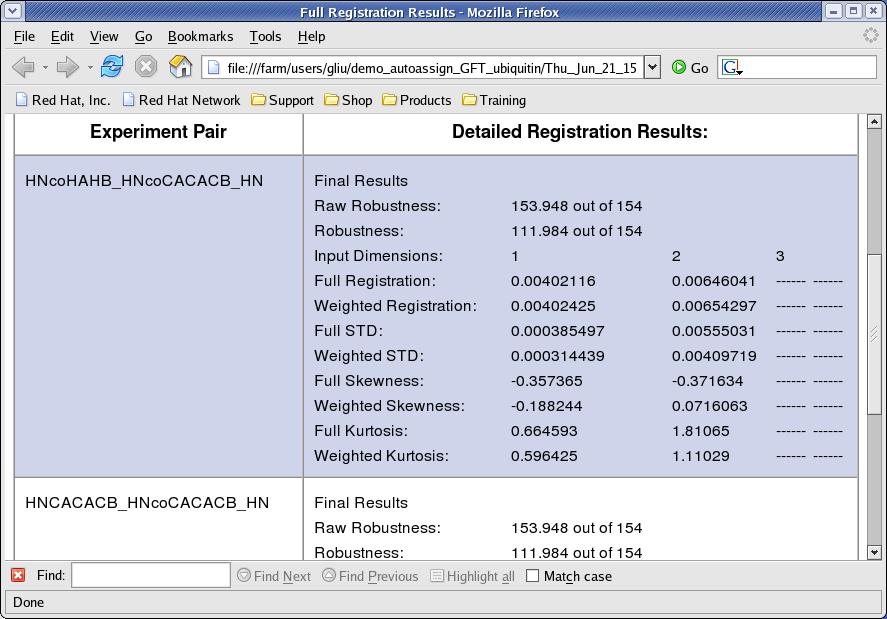
- View Registration [button]: the interface will create an html page by consolidating .reg files generated and will try to open it with a browser. The
Weighted Registrationvalues are used to register peak lists. TheFull Stdvalues are used in calculating tolerances included in the control file (Figure 5). - Revert to Original Value [button]: this function refills text fields for registration value with original registration value calculated.
- Apply Registration [button]: this function applies registration values to each peaklist. It creates new peaklist files with _shifted.pks extension.
Figure 5: Registration Results Page
Dataset Properties
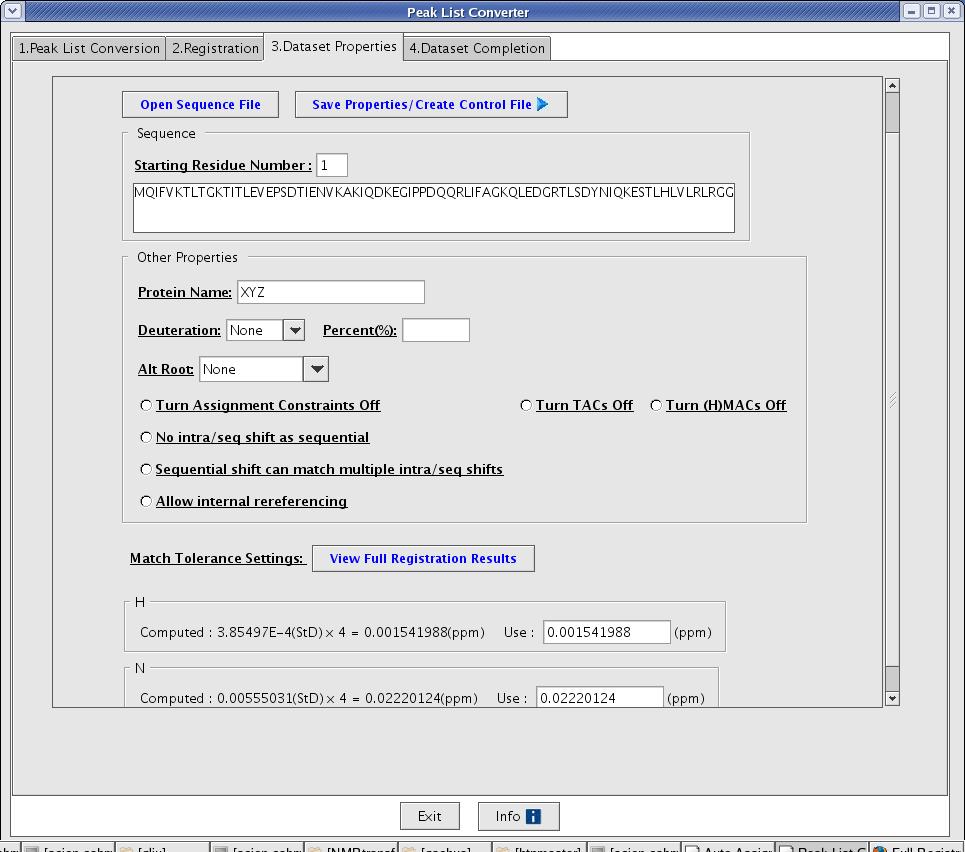
After registration, the converter moves to the Data Set Properties page (Figure 6). A user is asked to provide additional information to complete the control file creation. The value entered by a user on this tab will be used in the properties, tolerances and sequence sections in a control file.
- Open Sequence File [button]: this function opens a window for a user to select a sequence file.
- Save Properties/Create Control File [button]: this function saves dataset property information into an internal data structure and use it to produce a control file. This button should be clicked to produce an initial control file and the result will be displayed on the next tab.
Figure 6: Data Set Properties Page
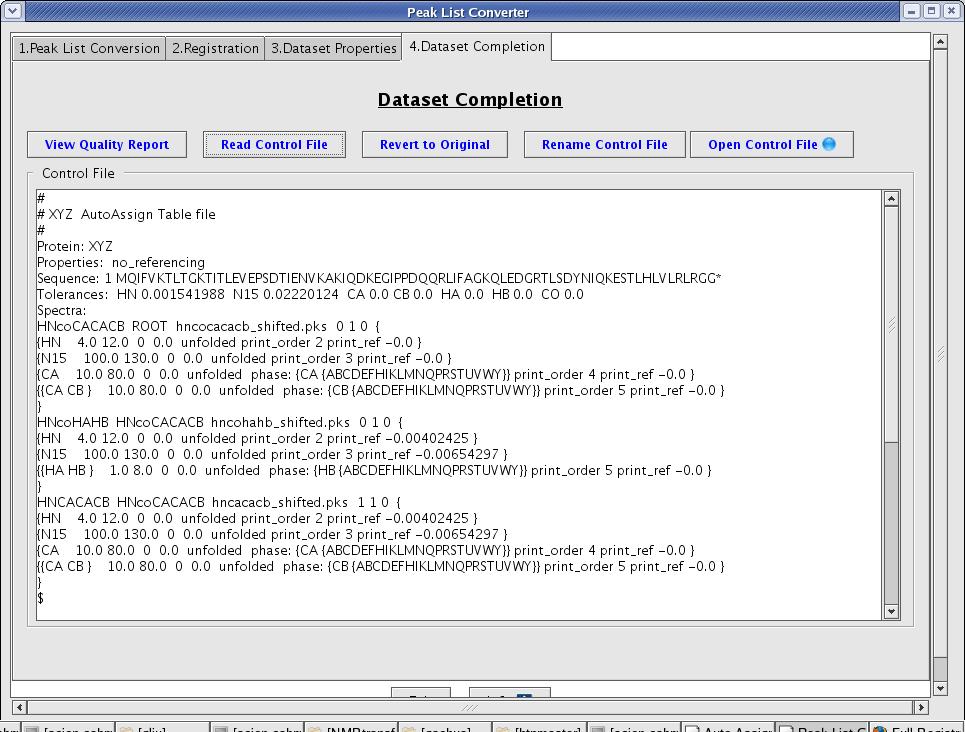
Dataset Completion
A user can review the control file generated by the interface, manually edit its content, and then save it (Figure 7). To save a revised control file select the Rename Control File button.
- View Quality Report [button]: this function opens a window for a user to view quality assessment report on the dataset.
- Read Control File [button]: this function allows a user to import an existing control file to view.
- Revert to Original [button]: this function brings back original control file to the screen.
- Rename Control File [button]: this function allows to save a control file with a different name.
- Open Control File [button]: this function opens a control file for analysis. The default control file is the one with original values. When a user saved the control file, that file becomes the default.
Figure 7: Peak List Converter Page
Follow the Dataset Creation process that the interface guides you through. At the end, you will have a dataset in AutoAssign format. Hitting the Open Control File button launches a new AutoAssign connectivity map window, and the user can proceed to computing resonance assignments.
Manually Preparation of Peak Files (UB)
One can manually prepare input files by using the follow format, UBNMR can also be used to prepare the input files for AutoAssign. In UBNMR run makeAutoList to generate input files for AutoAssign.
- Peak File Format:
<Index> <dim1> <dim2> [dimX] <Intensity> <Label>[.PeakNotes] ... *
- Here is an example HNCA peak list file:
#Index Xppm Yppm Zppm Intensity Label.notes
126 8.871 110.859 50.247 3242120 HNCA
125 8.870 110.898 62.529 724463 HNCA.;g45
73 8.744 116.161 56.614 2287600 HNCA
:
:
145 9.153 112.004 57.415 2788050 HNCA
where individual fields are separated by spaces and/or tabs, and comments are indicated by a "#" sign at the beginning of a line.
Performing Resonance Assignments
Connect to the server and open control-file
- One can open a dataset to analyze start from the main menu by selecting
File+ -> =Open Control Filemenu option or pressing theOpen Control Filebutton on the 4th tab (dataset completion) of the Dataset Creation Graphical User Interface. - AutoAssign will bring up the following window prompting the user to connect to a server (Figure 8):
Figure 8: Connection to Server
If the client and server are on the same computer, you should be able to simply select "Connect".
- Next a blank
Connectivity Mapwindow is opened (Figure 9).
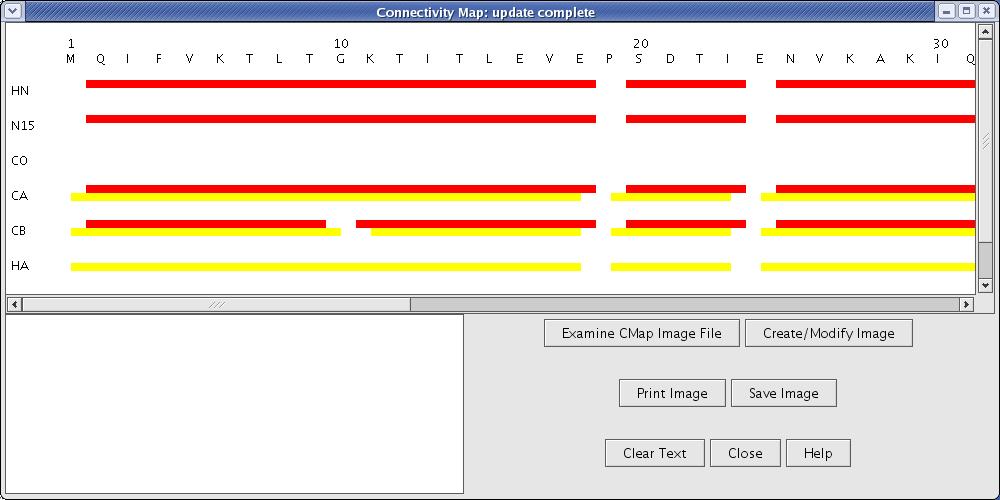
Figure 9: Connectivity Map
Starting a Backbone Resonance Assignment
Select the Execute -> Default Execution menu option to start the analysis.
Default Executionis the suggested method to run the program. TheRefined ExecutionExecute menu option run AutoAssign in an aggressive mode which is not as safe as theDefault Executionoption.- The
Connectivity Mapwill show the resonance assignment results as the analysis progresses. The whole analysis should only take a few seconds (Figure 10).
Figure 10: Assigned Connectivity Map
Assignment Constraints
The user can specify a variety of assignment constraints in the Notes column of a Sparky peak list using a controlled vocabulary. These assignment constraints include: spin system typing, grouping and mapping constraints which are used by AutoAssign in the assignment process. A complete description of assignment constraints and their usage in AutoAssign is provided in the online documentation.
Examining and Saving the Resonance Assignment Results
- The saving format options can be selected under the
save->Chemical Shiftsubmenus. - The
Specific GS,Unassigned Residues, andUnassigned GSsExamine menu options are very valuable to troubleshoot problems with assignments or missing assignments. These can be as guides to reexamine spectra to find missing peaks or to apply peak-based assignment constraints if such interventions are necessary. - The results file saved from
Examine->All GSsis used to convert the AutoAssign result to XEASY. - For individually assigned peak lists in Sparky format click:
Save->All Sparky Peaklists. - There are several options under
Save->Chemical Shifts for saving the assignments in bmrb formats (2.1 and 3.0) and connectivity map format (cmap).<span style="font-family: sans-serif;" /><span style="font-family: sans-serif;" />
The Connectivity Map Image Editor
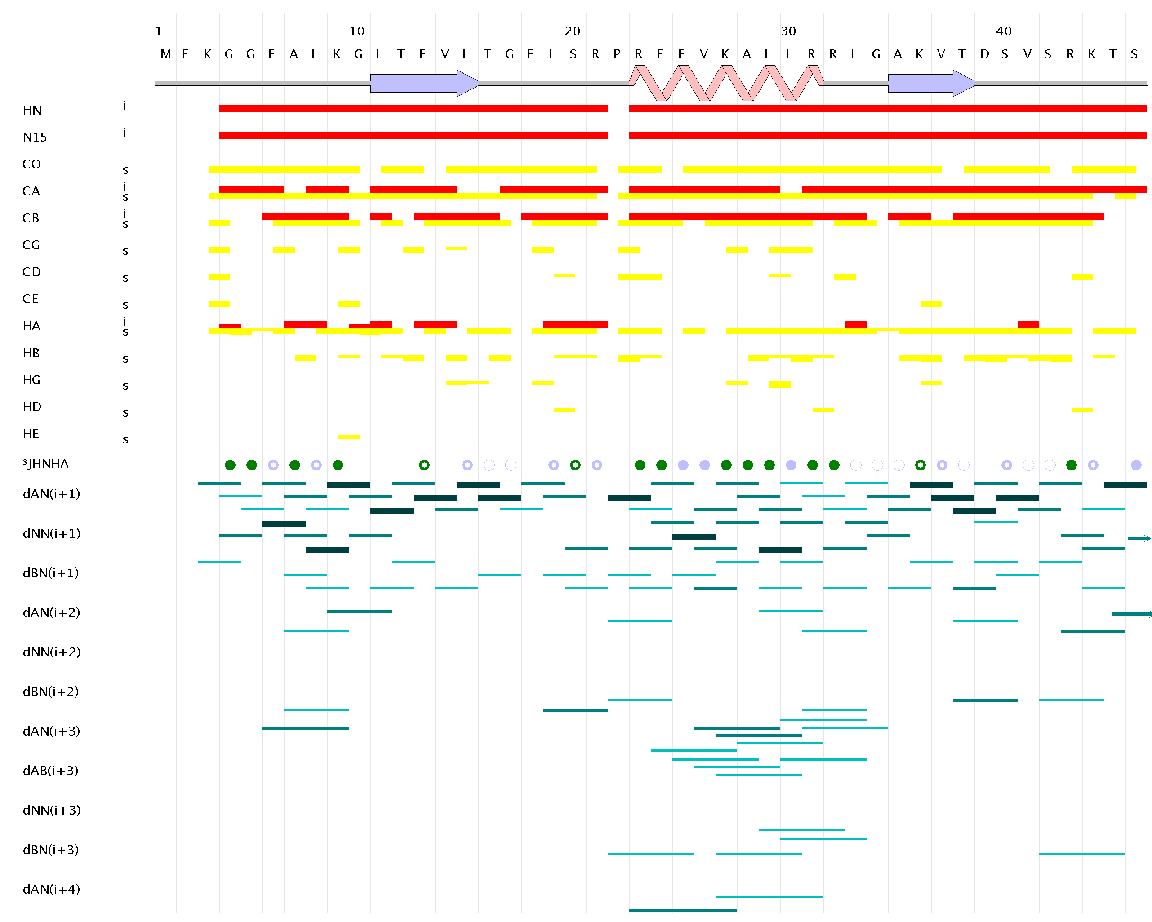
A connectivity map image represents resonance and other experimental data on the protein sequence in a graphical form. The CMap Image Editor (which can be launched under the Tools tab) allows one to create such an image (Figure 11). This facility is particularly useful for generating cmap images for publication. For a complete description of this editor and all of its functions see the on-line documentation.
Figure 11: A CMap Image
AutoPeak Scripts
The AutoAssign bin directory features a plethora of perl scripts written by Hunter Moseley for many functions related to AutoAssign and its interplay with various programs including Sparky, as well as interconversion between a variety of formats. A glossary of these scripts is available and a more in-depth description of the more commonly used scripts in our structure determination process is available here. Included among the major applications of the AutoPeak scripts is the creation and manipulation of chemical shift files in BMRB format. Note that AutoPeak scripts in AutoAssign versions above 1.15.1 are written for BMRB 3.0 format. Here is a list of the scripts we most often use in our structure determinations:
- bmrb2cmap.pl - converts bmrb format to cmap format. bmrbACS2SparkyPks.pl - converts bmrb assigned chemical shift save frame to Assigned Sparky Peak List format.
- bmrb2AutoStructure.pl - converts bmrb format to AutoStructure Resonance List format.
- compare_bmrb.pl - compares two bmrb files and returns how well they match.
- missing_shifts.pl - reports the missing AtomTypes in a given BMRB file.
- sparkyPks2bmrb.pl - converts a sparky peaklist into a BMRB assigned peak list save frame.
- sparkyRL2bmrb.pl- converts Sparky resonance list into a BMRB file.
References
1. Zimmerman, D.E., Kulikowski, C.A., Huang, Y., Feng, W., Tashiro, M., Shimotakahara, S., Chien, C., Powers, R. and Montelione, G.T. (1997) Automated analysis of protein NMR assignments using methods from artificial intelligence. J. Mol. Biol. 269, 592-610.
2. Moseley, H.N.B., Monleon, D. and Montelione, G.T. (2001) Automatic Determination of Protein Backbone Resonance Assignments from Triple Resonance NMR Data. Methods Enzymology 339, 91-108.
-- GaohuaLiu - 11 Jun 2007
-- Updated/Edited by JimAramini - 09 Nov 2009