AutoStructure: Difference between revisions
No edit summary |
No edit summary |
||
| Line 44: | Line 44: | ||
==== Other input files ==== | ==== Other input files ==== | ||
Other files that can be included in an AutoStructure calculation inlcude: | Other files that can be included in an AutoStructure calculation inlcude: | ||
*<sup>H</sup>N-H<sup>α</sup> J-couplings: used for initial secondary structrue anlysis | *<sup>H</sup>N-H<sup>α</sup> J-couplings: used for initial secondary structrue anlysis | ||
*slow exchanging NH list: used for initial secondary structrue anlysis | *slow exchanging NH list: used for initial secondary structrue anlysis | ||
*dihedral angle constraints: used directly in CYANA/XPLOR structure calculations | *dihedral angle constraints: used directly in CYANA/XPLOR structure calculations | ||
*maunal distance constraints: used directly in CYANA/XPLOR structure calculations | *maunal distance constraints: used directly in CYANA/XPLOR structure calculations | ||
=== Graphical User Interface === | === Graphical User Interface === | ||
AutoStructure features a GUI which is particularly useful for preparing the control file for and subsequent analysis of structure calulations. | AutoStructure features a GUI which is particularly useful for preparing the control file for and subsequent analysis of structure calulations. | ||
The main page (Figure 1) features the following pull-down menus: | The main page (Figure 1) features the following pull-down menus: | ||
*File: manipulation of control file | *File: manipulation of control file | ||
*AutoStructure: starting an AutoStructure run and analysis of an output directory | *AutoStructure: starting an AutoStructure run and analysis of an output directory | ||
*AutoQF(RPF): starting an RPF analysis and analysis of an RPF directory | *AutoQF(RPF): starting an RPF analysis and analysis of an RPF directory | ||
*PDB Tools: tools for converting PDB coordinates to IUPAC format | *PDB Tools: tools for converting PDB coordinates to IUPAC format | ||
*Misc Tools | *Misc Tools | ||
==== Figure 1: AutoStructure main page ==== | ==== Figure 1: AutoStructure main page ==== | ||
=== Control File<br> === | === Control File<br> === | ||
=== | === Starting a Calculation === | ||
The user can start a calculation from either the graphical user face or the command line. | The user can start a calculation from either the graphical user face or the command line. | ||
| Line 109: | Line 107: | ||
-v Calculate the M score and average shifts | -v Calculate the M score and average shifts | ||
</pre> | </pre> | ||
== '''References''' == | == '''References''' == | ||
-- JimAramini - 07 Nov 2009 | -- JimAramini - 07 Nov 2009 | ||
Revision as of 21:12, 7 November 2009
Introduction
Getting Started
Input Files
The following files are required to perform an AutoStructure run.
Protein sequence file
The sequence file must be in the following format:
1 @ MET 2 @ GLU 3 @ PHE 4 @ PRO 5 @ ASP 6 @ LEU 7 @ THR 8 @ VAL 9 @ GLU 10 @ ILE ....etc.
Here is an example sequence file.
Chemical shift assignment file
The chemical shift assignments for the protein must be in BMRB 2.1 format. The header information is ignored.
AutoSructure does interpret the ambiguity code column. This is important for denoting stereospecific assignments.
1 1 Met HA H 3.999 . 1 2 1 Met HB2 H 2.011 . 2 3 1 Met HB3 H 1.946 . 2 4 1 Met HG2 H 2.368 . 2 5 1 Met HG3 H 2.274 . 2 6 1 Met CA C 55.110 . 1 7 1 Met CB C 33.147 . 1 8 1 Met CG C 30.951 . 1 9 2 Glu HA H 4.548 . 1 ....etc.
Here is an example bmrb file.
NOESY peak lists
AutoStructure can accept 3D and 4D NOESY peak lists. In principle, any column formated peak list file can be read by the program; we typically use either Sparky or Xeasy formats.
Here are example 15N-edited NOESY and 13C-edited NOESY peak lists.
The column definitions and tolerances are defined in the control file (see below).
Other input files
Other files that can be included in an AutoStructure calculation inlcude:
- HN-Hα J-couplings: used for initial secondary structrue anlysis
- slow exchanging NH list: used for initial secondary structrue anlysis
- dihedral angle constraints: used directly in CYANA/XPLOR structure calculations
- maunal distance constraints: used directly in CYANA/XPLOR structure calculations
Graphical User Interface
AutoStructure features a GUI which is particularly useful for preparing the control file for and subsequent analysis of structure calulations.
The main page (Figure 1) features the following pull-down menus:
- File: manipulation of control file
- AutoStructure: starting an AutoStructure run and analysis of an output directory
- AutoQF(RPF): starting an RPF analysis and analysis of an RPF directory
- PDB Tools: tools for converting PDB coordinates to IUPAC format
- Misc Tools
Figure 1: AutoStructure main page
Control File
Starting a Calculation
The user can start a calculation from either the graphical user face or the command line.
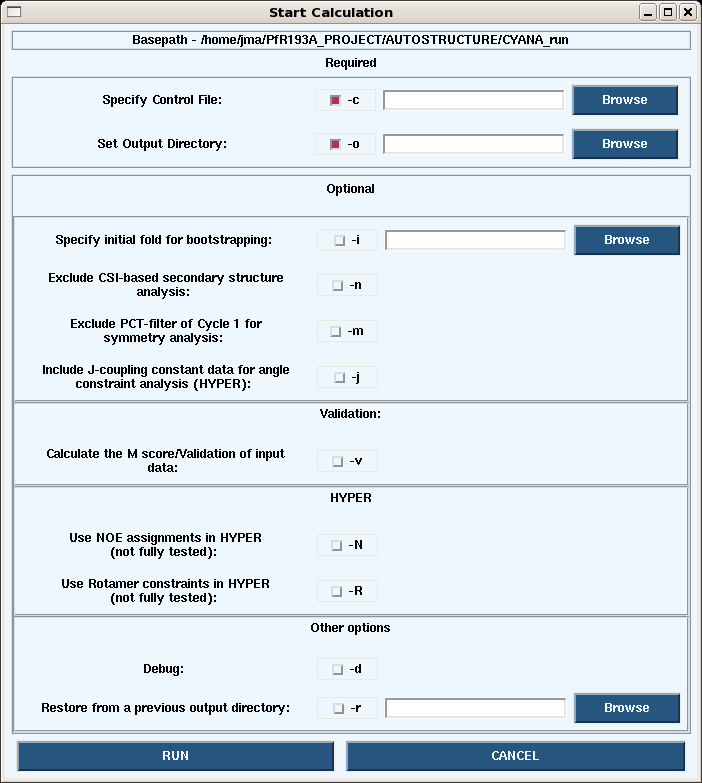
1. Launching a calculation from the GUI.
- Under the Autostructure pull-down on the main page, choosing Start -> Calc opens the dialogue box below.
- Launching calculations from the GUI uses is generally slow since only the processors on your own machine are used. To speed up the calculations use the command line on a cluster.
2. Launching a calculation from the command line.
- Login in the a cluster. At CABM we use AutoStructure version 2.2.1 on hummer.
- A simple command line run can be started as follows:
/farm/software/AutoStructure/AutoStructure-2.2.1/bin/autostructure -c controlfile_CYANArun -o testCYANArun.out -v
- Running the autostructure command gives the following options (like those avaliable in the GUI):
AutoStructure/RPF Version 2.2.1 Copyright(C) 2007
Center for Advanced Biotechnology and Medicine (CABM)
Rutgers University
Options:
-c control_file Required
-o output_dir Required
-d For debug
-h Help
-m Exclude PCT-filter of Cycle1 for symmetry analysis
-n Exclude CSI-based secondary structure analysis
-i structure_file inital fold for bootstrapping
-j Include J-coupling constant data for angle constraint analysis (HYPER)
-k float_number Calibration coefficient
-N Include NOE assignments in HYPER caluclation (under development)
-q structure_file AutoQF-Calculate the F and DP scores of the input structure_file (IUPAC naming)
-r path Restore from a prior outout_dir
-R Include rotamer constraints in HYPER calculation (under development)
-v Calculate the M score and average shifts
References
-- JimAramini - 07 Nov 2009