Resonance Assignment/CARA: Difference between revisions
No edit summary |
|||
| Line 1: | Line 1: | ||
== Loading a | == Loading a new template == | ||
To start a new structure determination project in CARA you need to load a template. A template is a CARA repository without project that contains definitions for residue types, spectrum type and LUA scripts. You can either load the default template from the [http://www.cara.ethz.ch/Wiki/TemplatesPage templates page] of the official CARA web-site, or extract a template from an existing CARA repository. | To start a new structure determination project in CARA you need to load a template. A template is a CARA repository without project that contains definitions for residue types, spectrum type and LUA scripts. You can either load the default template from the [http://www.cara.ethz.ch/Wiki/TemplatesPage templates page] of the official CARA web-site, or extract a template from an existing CARA repository. | ||
| Line 8: | Line 8: | ||
CARA Template for GFT spectra [[Media:BoR54_template.cara|BoR54_template.cara]]: Latest GFT template for BoR54 project | CARA Template for GFT spectra [[Media:BoR54_template.cara|BoR54_template.cara]]: Latest GFT template for BoR54 project | ||
== Starting a new project == | |||
Normally you would create a new project based on a protein sequence in XEASY format. | |||
If you have the protein sequence in one-letter format you can convert it to XEASY <tt>.seq</tt> file the following way: | |||
#Save the one-letter sequence in a <tt>.aa</tt> file, e.g. <tt>foo.aa</tt> | |||
#Execute '''OneLetterFileToSeqFile''' LUA script from '''Terminal''' tab of the main window. Select your <tt>.aa</tt> file as input and give full path for the output <tt>.seq</tt> file. | |||
[[Image:CARA One Letter to Seqfile.png]] | |||
Revision as of 23:57, 3 December 2009
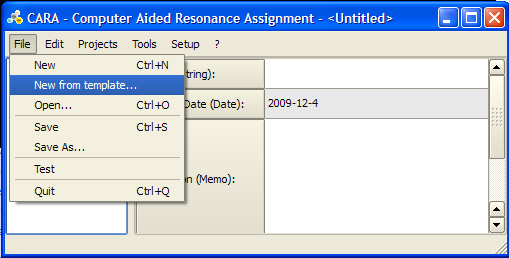
Loading a new template
To start a new structure determination project in CARA you need to load a template. A template is a CARA repository without project that contains definitions for residue types, spectrum type and LUA scripts. You can either load the default template from the templates page of the official CARA web-site, or extract a template from an existing CARA repository.
In CARA click File -> New from template and select the appropriate .cara file. For detailed instructions see http://www.cara.ethz.ch/Wiki/ImportingTemplate
CARA Template for GFT spectra BoR54_template.cara: Latest GFT template for BoR54 project
Starting a new project
Normally you would create a new project based on a protein sequence in XEASY format.
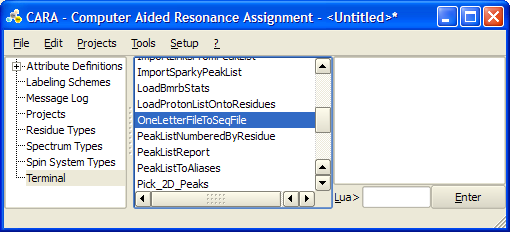
If you have the protein sequence in one-letter format you can convert it to XEASY .seq file the following way:
- Save the one-letter sequence in a .aa file, e.g. foo.aa
- Execute OneLetterFileToSeqFile LUA script from Terminal tab of the main window. Select your .aa file as input and give full path for the output .seq file.