Paramagnetic Constraints in Structure Determination: Difference between revisions
No edit summary |
No edit summary |
||
| Line 933: | Line 933: | ||
== '''References''' == | == '''References''' == | ||
1. Liu, Y.Z., and Prestegard, J.H. (2008) Direct measurement of dipole-dipole/CSA cross-correlated relaxation by a constant-time experiment. ''J. Magn. Reson. 193'', 23-31. | [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2542487/?tool=pubmed 1. Liu, Y.Z., and Prestegard, J.H. (2008) Direct measurement of dipole-dipole/CSA cross-correlated relaxation by a constant-time experiment. ''J. Magn. Reson. 193'', 23-31.] | ||
2. Schwieters, C.D., Kuszewski, J.J., and Clore, G.M. (2006). Using Xplor-NIH for NMR molecular structure determination. ''Prog. NMR Spect. 48'', 47-62.<br> | 2. Schwieters, C.D., Kuszewski, J.J., and Clore, G.M. (2006). Using Xplor-NIH for NMR molecular structure determination. ''Prog. NMR Spect. 48'', 47-62.<br> | ||
3. Banci, L., Bertini, I., Cavallaro, G., Giachetti, A., Luchinat, C., and Parigi, G. (2004) Paramagnetism-based restraints for Xplor-NIH. ''J. Biomol. NMR 28'', 249-261. | [http://www.ncbi.nlm.nih.gov/pubmed/14752258?itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum&ordinalpos=2 3. Banci, L., Bertini, I., Cavallaro, G., Giachetti, A., Luchinat, C., and Parigi, G. (2004) Paramagnetism-based restraints for Xplor-NIH. ''J. Biomol. NMR 28'', 249-261.] | ||
4. Valafar, H., and Prestegard, J.H. (2004) REDCAT: a residual dipolar coupling analysis tool. ''J. Magn. Reson. 167'', 228-241. | [http://www.ncbi.nlm.nih.gov/pubmed/15040978?itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum&ordinalpos=6 4. Valafar, H., and Prestegard, J.H. (2004) REDCAT: a residual dipolar coupling analysis tool. ''J. Magn. Reson. 167'', 228-241.] | ||
5. <font face="Arial">Iwahara, J., Schwieters, C.D., and Clore, G.M. (2004) Ensemble approach for NMR structure refinement against | [http://www.ncbi.nlm.nih.gov/pubmed/15125681?itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum&ordinalpos=2 5. <font face="Arial">Iwahara, J., Schwieters, C.D., and Clore, G.M. (2004) Ensemble approach for NMR structure refinement against (1)H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. ''J Am Chem Soc''. ''126'', 5879-96.</font>] | ||
Revision as of 16:48, 5 January 2010
Introduction
The collection of paramagnetic constraint data and the use of those data as a part of a structure determination is fairly straight forward. RDCs can be collected using either the J-modulation or HSQC-TROSY methods described in the RDC section. Paramagnetic relaxation enhancements (PREs) are collected based on attenuation of signal intensity in HSQC or TROSY spectra. When Curie contributions from paramagnetic metals dominate it will be convenient to make measurements at multiple field strengths as the effects are field squared dependent and field variation provides a useful way to probe different distance ranges. We have access to spectrometers operating from 600 to 900 MHz. Pseudocontact shifts (PCSs) are measured by comparison of HSQC (or TROSY) cross-peak positions in diamagnetic (La3+) and paramagnetic complexes (Dy3+ or Tb3+). Pairing of shifted and non-shifted peaks is facilitated by the fact that shifts in in both 1H and 15N dimensions are nearly equal on the ppm scale and are therefore connected by diagonal lines. Paramagnetic relaxation interferences (PRIs) produce differential effects on the α and β cross peaks of coupled HSQC spectra and the cross-correlation effects can be measured using experiments that we have developed for the measurement of correlation times from CSA/DD interference [1]. Integration of these data into structure characterization protocols in the NESG is accomplished using programs such as XPLOR-NIH [2,3] or REDCAT [4]. CYANA can also accommodate pseudocontact shifts.
Protocols
Generation of MTSL-cysteine pseudo-residue
Editing the Xplor/CNS topology library
Apply the following lines to protein-allhdg.top:
residue CYSM
group
atom N type=NH1 charge=-0.36 end
atom HN type=H charge= 0.26 end
atom CA type=CH1E charge= 0.00 end
atom HA type=HA charge= 0.10 end
atom CB type=CH2E charge=-0.20 end
atom HB1 type=HA charge= 0.10 end
atom HB2 type=HA charge= 0.10 end
atom SG type=SH1E charge=-0.05 end
! atom HG type=H charge= 0.05 end
atom C type=C charge= 0.48 end
atom O type=O charge=-0.48 end
ATOM CAE TYPE= CMAE CHARGE= 0.016 END
ATOM CAL TYPE= CMAL CHARGE= 0.097 END
ATOM CAF TYPE= CMAF CHARGE= 0.016 END
ATOM NAI TYPE= NMAI CHARGE=-0.164 END
ATOM OAB TYPE= OMAB CHARGE=-0.114 END
! ATOM HAA TYPE= MMAA CHARGE= 0.029 END
ATOM CAK TYPE= CMAK CHARGE= 0.097 END
ATOM CAC TYPE= CMAC CHARGE= 0.016 END
ATOM CAD TYPE= CMAD CHARGE= 0.016 END
ATOM CAG TYPE= CMAG CHARGE=-0.042 END
ATOM CAJ TYPE= CMAJ CHARGE=-0.011 END
ATOM CAH TYPE= CMAH CHARGE= 0.038 END
ATOM SAA TYPE= SMAA CHARGE= 0.041 END
ATOM HAG TYPE = MMA charge= 0.14 end !yizhou
ATOM HAH1 type=MMA charge= 0.10 end
ATOM HAH2 type=MMA charge= 0.10 end
ATOM HAE1 type=MMA charge= 0.10 end
ATOM HAE2 type=MMA charge= 0.10 end
ATOM HAE3 type=MMA charge= 0.10 end
ATOM HAF1 type=MMA charge= 0.10 end
ATOM HAF2 type=MMA charge= 0.10 end
ATOM HAF3 type=MMA charge= 0.10 end
ATOM HAD1 type=MMA charge= 0.10 end
ATOM HAD2 type=MMA charge= 0.10 end
ATOM HAD3 type=MMA charge= 0.10 end
ATOM HAC1 type=MMA charge= 0.10 end
ATOM HAC2 type=MMA charge= 0.10 end
ATOM HAC3 type=MMA charge= 0.10 end
bond N HN
bond N CA bond CA HA
bond CA CB bond CB HB1 bond CB HB2
bond CB SG ! bond SG HG
bond CA C
bond C O
bond SG SAA !yizhou
bond CAG HAG !yizhou
bond CAH HAH1
bond CAH HAH2
bond CAE HAE1
bond CAE HAE2
bond CAE HAE3
bond CAF HAF1
bond CAF HAF2
bond CAF HAF3
bond CAC HAC1
bond CAC HAC2
bond CAC HAC3
bond CAD HAD1
bond CAD HAD2
bond CAD HAD3
BOND CAE CAL
BOND CAL CAF
BOND CAL NAI
BOND CAL CAJ
BOND NAI OAB
BOND NAI CAK
! BOND OAB HAA
BOND CAK CAC
BOND CAK CAD
BOND CAK CAG
BOND CAG CAJ
BOND CAJ CAH
BOND CAH SAA
ANGLE CAE CAL CAF
ANGLE CAE CAL NAI
ANGLE CAE CAL CAJ
ANGLE CAF CAL NAI
ANGLE CAF CAL CAJ
ANGLE NAI CAL CAJ
ANGLE CAL NAI OAB
ANGLE CAL NAI CAK
ANGLE OAB NAI CAK
! ANGLE NAI OAB HAA
ANGLE NAI CAK CAC
ANGLE NAI CAK CAD
ANGLE NAI CAK CAG
ANGLE CAC CAK CAD
ANGLE CAC CAK CAG
ANGLE CAD CAK CAG
ANGLE CAK CAG CAJ
ANGLE CAL CAJ CAG
ANGLE CAL CAJ CAH
ANGLE CAG CAJ CAH
ANGLE CAJ CAH SAA
ANGLE CAH SAA SG
angle SAA SG CB !yizhou
angle HAG CAG CAK !yizhou
angle HAG CAG CAJ !yizhou
! IMPROPER CAJ CAL CAG CAH
! IMPROPER CAL CAE CAF NAI
! IMPROPER CAK NAI CAD CAC
IMPROPER NAI CAL OAB CAK
IMPROPER HAH1 HAH2 SAA CAJ
IMPROPER HAE1 HAE2 CAL HAE3
IMPROPER HAF1 HAF2 CAL HAF3
IMPROPER HAC1 HAC2 CAK HAC3
IMPROPER HAD1 HAD2 CAK HAD3
! DIHEDRAL CAE CAL NAI CAK
! DIHEDRAL CAE CAL CAJ CAH
! DIHEDRAL CAL NAI OAB HAA
! DIHEDRAL CAG CAK NAI CAL
! DIHEDRAL NAI CAK CAG CAJ
! DIHEDRAL CAK CAG CAJ CAH
DIHEDRAL CAL CAJ CAH SAA
! DIHEDRAL SG SAA CAH CAJ
! dihedral OAB NAI CAL CAJ
dihedral HAG CAG CAJ CAL !yizhou
improper HA N C CB !chirality CA
improper HB1 HB2 CA SG !stereo CB
dihedral SG CB CA N
end
residue C2M
group
atom N type=NH1 charge=-0.36 end
atom HN type=H charge= 0.26 end
atom CA type=CH1E charge= 0.00 end
atom HA type=HA charge= 0.10 end
atom CB type=CH2E charge=-0.20 end
atom HB1 type=HA charge= 0.10 end
atom HB2 type=HA charge= 0.10 end
atom SG type=SH1E charge=-0.05 end
! atom HG type=H charge= 0.05 end
atom C type=C charge= 0.48 end
atom O type=O charge=-0.48 end
ATOM CAE TYPE= CMAE CHARGE= 0.016 END
ATOM CAL TYPE= CMAL CHARGE= 0.097 END
ATOM CAF TYPE= CMAF CHARGE= 0.016 END
ATOM NAI TYPE= NMAI CHARGE=-0.164 END
ATOM OAB TYPE= OMAB CHARGE=-0.114 END
! ATOM HAA TYPE= MMAA CHARGE= 0.029 END
ATOM CAK TYPE= CMAK CHARGE= 0.097 END
ATOM CAC TYPE= CMAC CHARGE= 0.016 END
ATOM CAD TYPE= CMAD CHARGE= 0.016 END
ATOM CAG TYPE= CMAG CHARGE=-0.042 END
ATOM CAJ TYPE= CMAJ CHARGE=-0.011 END
ATOM CAH TYPE= CMAH CHARGE= 0.038 END
ATOM SAA TYPE= SMAA CHARGE= 0.041 END
ATOM CBE TYPE= CMAE CHARGE= 0.016 END
ATOM CBL TYPE= CMAL CHARGE= 0.097 END
ATOM CBF TYPE= CMAF CHARGE= 0.016 END
ATOM NBI TYPE= NMAI CHARGE=-0.164 END
ATOM OBB TYPE= OMAB CHARGE=-0.114 END
ATOM CBK TYPE= CMAK CHARGE= 0.097 END
ATOM CBC TYPE= CMAC CHARGE= 0.016 END
ATOM CBD TYPE= CMAD CHARGE= 0.016 END
ATOM CBG TYPE= CMAG CHARGE=-0.042 END
ATOM CBJ TYPE= CMAJ CHARGE=-0.011 END
ATOM CBH TYPE= CMAH CHARGE= 0.038 END
ATOM SBA TYPE= SMAA CHARGE= 0.041 END
! ATOM SBG type=SM1E charge=-0.05 end
ATOM HAG TYPE = MMA charge= 0.14 end !yizhou
ATOM HAH1 type=MMA charge= 0.10 end
ATOM HAH2 type=MMA charge= 0.10 end
ATOM HAE1 type=MMA charge= 0.10 end
ATOM HAE2 type=MMA charge= 0.10 end
ATOM HAE3 type=MMA charge= 0.10 end
ATOM HAF1 type=MMA charge= 0.10 end
ATOM HAF2 type=MMA charge= 0.10 end
ATOM HAF3 type=MMA charge= 0.10 end
ATOM HAD1 type=MMA charge= 0.10 end
ATOM HAD2 type=MMA charge= 0.10 end
ATOM HAD3 type=MMA charge= 0.10 end
ATOM HAC1 type=MMA charge= 0.10 end
ATOM HAC2 type=MMA charge= 0.10 end
ATOM HAC3 type=MMA charge= 0.10 end
ATOM HBG TYPE = MMA charge= 0.14 end !yizhou
ATOM HBH1 type=MMA charge= 0.10 end
ATOM HBH2 type=MMA charge= 0.10 end
ATOM HBE1 type=MMA charge= 0.10 end
ATOM HBE2 type=MMA charge= 0.10 end
ATOM HBE3 type=MMA charge= 0.10 end
ATOM HBF1 type=MMA charge= 0.10 end
ATOM HBF2 type=MMA charge= 0.10 end
ATOM HBF3 type=MMA charge= 0.10 end
ATOM HBD1 type=MMA charge= 0.10 end
ATOM HBD2 type=MMA charge= 0.10 end
ATOM HBD3 type=MMA charge= 0.10 end
ATOM HBC1 type=MMA charge= 0.10 end
ATOM HBC2 type=MMA charge= 0.10 end
ATOM HBC3 type=MMA charge= 0.10 end
bond N HN
bond N CA bond CA HA
bond CA CB bond CB HB1 bond CB HB2
bond CB SG ! bond SG HG
bond CA C
bond C O
bond SG SAA !yizhou
bond CAG HAG !yizhou
bond CAH HAH1
bond CAH HAH2
bond CAE HAE1
bond CAE HAE2
bond CAE HAE3
bond CAF HAF1
bond CAF HAF2
bond CAF HAF3
bond CAC HAC1
bond CAC HAC2
bond CAC HAC3
bond CAD HAD1
bond CAD HAD2
bond CAD HAD3
BOND CAE CAL
BOND CAL CAF
BOND CAL NAI
BOND CAL CAJ
BOND NAI OAB
BOND NAI CAK
! BOND OAB HAA
BOND CAK CAC
BOND CAK CAD
BOND CAK CAG
BOND CAG CAJ
BOND CAJ CAH
BOND CAH SAA
! bond CB SG ! bond SG HG
! bond SG SBA !yizhou
bond SG SBA
bond CBG HBG !yizhou
bond CBH HBH1
bond CBH HBH2
bond CBE HBE1
bond CBE HBE2
bond CBE HBE3
bond CBF HBF1
bond CBF HBF2
bond CBF HBF3
bond CBC HBC1
bond CBC HBC2
bond CBC HBC3
bond CBD HBD1
bond CBD HBD2
bond CBD HBD3
BOND CBE CBL
BOND CBL CBF
BOND CBL NBI
BOND CBL CBJ
BOND NBI OBB
BOND NBI CBK
! BOND OBB HBA
BOND CBK CBC
BOND CBK CBD
BOND CBK CBG
BOND CBG CBJ
BOND CBJ CBH
BOND CBH SBA
ANGLE CAE CAL CAF
ANGLE CAE CAL NAI
ANGLE CAE CAL CAJ
ANGLE CAF CAL NAI
ANGLE CAF CAL CAJ
ANGLE NAI CAL CAJ
ANGLE CAL NAI OAB
ANGLE CAL NAI CAK
ANGLE OAB NAI CAK
! ANGLE NAI OAB HAA
ANGLE NAI CAK CAC
ANGLE NAI CAK CAD
ANGLE NAI CAK CAG
ANGLE CAC CAK CAD
ANGLE CAC CAK CAG
ANGLE CAD CAK CAG
ANGLE CAK CAG CAJ
ANGLE CAL CAJ CAG
ANGLE CAL CAJ CAH
ANGLE CAG CAJ CAH
ANGLE CAJ CAH SAA
ANGLE CAH SAA SG
angle SAA SG CB !yizhou
angle HAG CAG CAK !yizhou
angle HAG CAG CAJ !yizhou
ANGLE CBE CBL CBF
ANGLE CBE CBL NBI
ANGLE CBE CBL CBJ
ANGLE CBF CBL NBI
ANGLE CBF CBL CBJ
ANGLE NBI CBL CBJ
ANGLE CBL NBI OBB
ANGLE CBL NBI CBK
ANGLE OBB NBI CBK
! ANGLE NBI OBB HBA
ANGLE NBI CBK CBC
ANGLE NBI CBK CBD
ANGLE NBI CBK CBG
ANGLE CBC CBK CBD
ANGLE CBC CBK CBG
ANGLE CBD CBK CBG
ANGLE CBK CBG CBJ
ANGLE CBL CBJ CBG
ANGLE CBL CBJ CBH
ANGLE CBG CBJ CBH
ANGLE CBJ CBH SBA
ANGLE CBH SBA SG
angle SBA SG CB !yizhou
angle HBG CBG CBK !yizhou
angle HBG CBG CBJ !yizhou
! IMPROPER CAJ CAL CAG CAH
! IMPROPER CAL CAE CAF NAI
! IMPROPER CAK NAI CAD CAC
IMPROPER NAI CAL OAB CAK
IMPROPER NBI CBL OBB CBK
IMPROPER HAH1 HAH2 SAA CAJ
IMPROPER HAE1 HAE2 CAL HAE3
IMPROPER HAF1 HAF2 CAL HAF3
IMPROPER HAC1 HAC2 CAK HAC3
IMPROPER HAD1 HAD2 CAK HAD3
IMPROPER HBH1 HBH2 SBA CBJ
IMPROPER HBE1 HBE2 CBL HBE3
IMPROPER HBF1 HBF2 CBL HBF3
IMPROPER HBC1 HBC2 CBK HBC3
IMPROPER HBD1 HBD2 CBK HBD3
! DIHEDRAL CAE CAL NAI CAK
! DIHEDRAL CAE CAL CAJ CAH
! DIHEDRAL CAL NAI OAB HAA
! DIHEDRAL CAG CAK NAI CAL
! DIHEDRAL NAI CAK CAG CAJ
! DIHEDRAL CAK CAG CAJ CAH
DIHEDRAL CAL CAJ CAH SAA
! DIHEDRAL SG SAA CAH CAJ
! dihedral OAB NAI CAL CAJ
dihedral HAG CAG CAJ CAL !yizhou
dihedral HBG CBG CBJ CBL !yizhou
improper HA N C CB !chirality CA
improper HB1 HB2 CA SG !stereo CB
dihedral SG CB CA N
end
residue C3M
group
atom N type=NH1 charge=-0.36 end
atom HN type=H charge= 0.26 end
atom CA type=CH1E charge= 0.00 end
atom HA type=HA charge= 0.10 end
atom CB type=CH2E charge=-0.20 end
atom HB1 type=HA charge= 0.10 end
atom HB2 type=HA charge= 0.10 end
atom SG type=SH1E charge=-0.05 end
! atom HG type=H charge= 0.05 end
atom C type=C charge= 0.48 end
atom O type=O charge=-0.48 end
ATOM CAE TYPE= CMAE CHARGE= 0.016 END
ATOM CAL TYPE= CMAL CHARGE= 0.097 END
ATOM CAF TYPE= CMAF CHARGE= 0.016 END
ATOM NAI TYPE= NMAI CHARGE=-0.164 END
ATOM OAB TYPE= OMAB CHARGE=-0.114 END
! ATOM HAA TYPE= MMAA CHARGE= 0.029 END
ATOM CAK TYPE= CMAK CHARGE= 0.097 END
ATOM CAC TYPE= CMAC CHARGE= 0.016 END
ATOM CAD TYPE= CMAD CHARGE= 0.016 END
ATOM CAG TYPE= CMAG CHARGE=-0.042 END
ATOM CAJ TYPE= CMAJ CHARGE=-0.011 END
ATOM CAH TYPE= CMAH CHARGE= 0.038 END
ATOM SAA TYPE= SMAA CHARGE= 0.041 END
ATOM CBE TYPE= CMAE CHARGE= 0.016 END
ATOM CBL TYPE= CMAL CHARGE= 0.097 END
ATOM CBF TYPE= CMAF CHARGE= 0.016 END
ATOM NBI TYPE= NMAI CHARGE=-0.164 END
ATOM OBB TYPE= OMAB CHARGE=-0.114 END
ATOM CBK TYPE= CMAK CHARGE= 0.097 END
ATOM CBC TYPE= CMAC CHARGE= 0.016 END
ATOM CBD TYPE= CMAD CHARGE= 0.016 END
ATOM CBG TYPE= CMAG CHARGE=-0.042 END
ATOM CBJ TYPE= CMAJ CHARGE=-0.011 END
ATOM CBH TYPE= CMAH CHARGE= 0.038 END
ATOM SBA TYPE= SMAA CHARGE= 0.041 END
! ATOM SBG type=SM1E charge=-0.05 end
ATOM CCE TYPE= CMAE CHARGE= 0.016 END
ATOM CCL TYPE= CMAL CHARGE= 0.097 END
ATOM CCF TYPE= CMAF CHARGE= 0.016 END
ATOM NCI TYPE= NMAI CHARGE=-0.164 END
ATOM OCB TYPE= OMAB CHARGE=-0.114 END
ATOM CCK TYPE= CMAK CHARGE= 0.097 END
ATOM CCC TYPE= CMAC CHARGE= 0.016 END
ATOM CCD TYPE= CMAD CHARGE= 0.016 END
ATOM CCG TYPE= CMAG CHARGE=-0.042 END
ATOM CCJ TYPE= CMAJ CHARGE=-0.011 END
ATOM CCH TYPE= CMAH CHARGE= 0.038 END
ATOM SCA TYPE= SMAA CHARGE= 0.041 END
! ATOM SCG type=SM1E charge=-0.05 end
ATOM HAG TYPE = MMA charge= 0.14 end !yizhou
ATOM HAH1 type=MMA charge= 0.10 end
ATOM HAH2 type=MMA charge= 0.10 end
ATOM HAE1 type=MMA charge= 0.10 end
ATOM HAE2 type=MMA charge= 0.10 end
ATOM HAE3 type=MMA charge= 0.10 end
ATOM HAF1 type=MMA charge= 0.10 end
ATOM HAF2 type=MMA charge= 0.10 end
ATOM HAF3 type=MMA charge= 0.10 end
ATOM HAD1 type=MMA charge= 0.10 end
ATOM HAD2 type=MMA charge= 0.10 end
ATOM HAD3 type=MMA charge= 0.10 end
ATOM HAC1 type=MMA charge= 0.10 end
ATOM HAC2 type=MMA charge= 0.10 end
ATOM HAC3 type=MMA charge= 0.10 end
ATOM HBG TYPE = MMA charge= 0.14 end !yizhou
ATOM HBH1 type=MMA charge= 0.10 end
ATOM HBH2 type=MMA charge= 0.10 end
ATOM HBE1 type=MMA charge= 0.10 end
ATOM HBE2 type=MMA charge= 0.10 end
ATOM HBE3 type=MMA charge= 0.10 end
ATOM HBF1 type=MMA charge= 0.10 end
ATOM HBF2 type=MMA charge= 0.10 end
ATOM HBF3 type=MMA charge= 0.10 end
ATOM HBD1 type=MMA charge= 0.10 end
ATOM HBD2 type=MMA charge= 0.10 end
ATOM HBD3 type=MMA charge= 0.10 end
ATOM HBC1 type=MMA charge= 0.10 end
ATOM HBC2 type=MMA charge= 0.10 end
ATOM HBC3 type=MMA charge= 0.10 end
ATOM HCG TYPE = MMA charge= 0.14 end !yizhou
ATOM HCH1 type=MMA charge= 0.10 end
ATOM HCH2 type=MMA charge= 0.10 end
ATOM HCE1 type=MMA charge= 0.10 end
ATOM HCE2 type=MMA charge= 0.10 end
ATOM HCE3 type=MMA charge= 0.10 end
ATOM HCF1 type=MMA charge= 0.10 end
ATOM HCF2 type=MMA charge= 0.10 end
ATOM HCF3 type=MMA charge= 0.10 end
ATOM HCD1 type=MMA charge= 0.10 end
ATOM HCD2 type=MMA charge= 0.10 end
ATOM HCD3 type=MMA charge= 0.10 end
ATOM HCC1 type=MMA charge= 0.10 end
ATOM HCC2 type=MMA charge= 0.10 end
ATOM HCC3 type=MMA charge= 0.10 end
bond N HN
bond N CA bond CA HA
bond CA CB bond CB HB1 bond CB HB2
bond CB SG ! bond SG HG
bond CA C
bond C O
bond SG SAA !yizhou
bond CAG HAG !yizhou
bond CAH HAH1
bond CAH HAH2
bond CAE HAE1
bond CAE HAE2
bond CAE HAE3
bond CAF HAF1
bond CAF HAF2
bond CAF HAF3
bond CAC HAC1
bond CAC HAC2
bond CAC HAC3
bond CAD HAD1
bond CAD HAD2
bond CAD HAD3
BOND CAE CAL
BOND CAL CAF
BOND CAL NAI
BOND CAL CAJ
BOND NAI OAB
BOND NAI CAK
! BOND OAB HAA
BOND CAK CAC
BOND CAK CAD
BOND CAK CAG
BOND CAG CAJ
BOND CAJ CAH
BOND CAH SAA
! bond CB SG ! bond SG HG
! bond SG SBA !yizhou
bond SG SBA
bond CBG HBG !yizhou
bond CBH HBH1
bond CBH HBH2
bond CBE HBE1
bond CBE HBE2
bond CBE HBE3
bond CBF HBF1
bond CBF HBF2
bond CBF HBF3
bond CBC HBC1
bond CBC HBC2
bond CBC HBC3
bond CBD HBD1
bond CBD HBD2
bond CBD HBD3
BOND CBE CBL
BOND CBL CBF
BOND CBL NBI
BOND CBL CBJ
BOND NBI OBB
BOND NBI CBK
! BOND OBB HBA
BOND CBK CBC
BOND CBK CBD
BOND CBK CBG
BOND CBG CBJ
BOND CBJ CBH
BOND CBH SBA
bond SG SCA
bond CCG HCG !yizhou
bond CCH HCH1
bond CCH HCH2
bond CCE HCE1
bond CCE HCE2
bond CCE HCE3
bond CCF HCF1
bond CCF HCF2
bond CCF HCF3
bond CCC HCC1
bond CCC HCC2
bond CCC HCC3
bond CCD HCD1
bond CCD HCD2
bond CCD HCD3
BOND CCE CCL
BOND CCL CCF
BOND CCL NCI
BOND CCL CCJ
BOND NCI OCB
BOND NCI CCK
! BOND OCB HCA
BOND CCK CCC
BOND CCK CCD
BOND CCK CCG
BOND CCG CCJ
BOND CCJ CCH
BOND CCH SCA
ANGLE CAE CAL CAF
ANGLE CAE CAL NAI
ANGLE CAE CAL CAJ
ANGLE CAF CAL NAI
ANGLE CAF CAL CAJ
ANGLE NAI CAL CAJ
ANGLE CAL NAI OAB
ANGLE CAL NAI CAK
ANGLE OAB NAI CAK
! ANGLE NAI OAB HAA
ANGLE NAI CAK CAC
ANGLE NAI CAK CAD
ANGLE NAI CAK CAG
ANGLE CAC CAK CAD
ANGLE CAC CAK CAG
ANGLE CAD CAK CAG
ANGLE CAK CAG CAJ
ANGLE CAL CAJ CAG
ANGLE CAL CAJ CAH
ANGLE CAG CAJ CAH
ANGLE CAJ CAH SAA
ANGLE CAH SAA SG
angle SAA SG CB !yizhou
angle HAG CAG CAK !yizhou
angle HAG CAG CAJ !yizhou
BOND CCK CCC
BOND CCK CCD
BOND CCK CCG
BOND CCG CCJ
BOND CCJ CCH
BOND CCH SCA
ANGLE CAE CAL CAF
ANGLE CAE CAL NAI
ANGLE CAE CAL CAJ
ANGLE CAF CAL NAI
ANGLE CAF CAL CAJ
ANGLE NAI CAL CAJ
ANGLE CAL NAI OAB
ANGLE CAL NAI CAK
ANGLE OAB NAI CAK
! ANGLE NAI OAB HAA
ANGLE NAI CAK CAC
ANGLE NAI CAK CAD
ANGLE NAI CAK CAG
ANGLE CAC CAK CAD
ANGLE CAC CAK CAG
ANGLE CAD CAK CAG
ANGLE CAK CAG CAJ
ANGLE CAL CAJ CAG
ANGLE CAL CAJ CAH
ANGLE CAG CAJ CAH
ANGLE CAJ CAH SAA
ANGLE CAH SAA SG
angle SAA SG CB !yizhou
angle HAG CAG CAK !yizhou
angle HAG CAG CAJ !yizhou
angle SAA SG SBA
angle SAA SG SCA
angle SBA SG SCA
! IMPROPER CAJ CAL CAG CAH
! IMPROPER CAL CAE CAF NAI
! IMPROPER CAK NAI CAD CAC
IMPROPER NAI CAL OAB CAK
IMPROPER NBI CBL OBB CBK
IMPROPER NCI CCL OCB CCK
IMPROPER HAH1 HAH2 SAA CAJ
IMPROPER HAE1 HAE2 CAL HAE3
IMPROPER HAF1 HAF2 CAL HAF3
IMPROPER HAC1 HAC2 CAK HAC3
IMPROPER HAD1 HAD2 CAK HAD3
IMPROPER HBH1 HBH2 SBA CBJ
IMPROPER HBE1 HBE2 CBL HBE3
IMPROPER HBF1 HBF2 CBL HBF3
IMPROPER HBC1 HBC2 CBK HBC3
IMPROPER HBD1 HBD2 CBK HBD3
IMPROPER HCH1 HCH2 SCA CCJ
IMPROPER HCE1 HCE2 CCL HCE3
IMPROPER HCF1 HCF2 CCL HCF3
IMPROPER HCC1 HCC2 CCK HCC3
IMPROPER HCD1 HCD2 CCK HCD3
! DIHEDRAL CAE CAL NAI CAK
! DIHEDRAL CAE CAL CAJ CAH
! DIHEDRAL CAL NAI OAB HAA
! DIHEDRAL CAG CAK NAI CAL
! DIHEDRAL NAI CAK CAG CAJ
! DIHEDRAL CAK CAG CAJ CAH
DIHEDRAL CAL CAJ CAH SAA
! DIHEDRAL SG SAA CAH CAJ
! dihedral OAB NAI CAL CAJ
dihedral HAG CAG CAJ CAL !yizhou
dihedral HBG CBG CBJ CBL !yizhou
dihedral HCG CCG CCJ CCL !yizhou
improper HA N C CB !chirality CA
improper HB1 HB2 CA SG !stereo CB
dihedral SG CB CA N
end
Apply the following lines to protein-allhdg.param
! param for MTSL
evaluate ($pd_x = 1.0)
eval ($pd_v=$pd_x* 16000.0) BOND CMAE CMAL $pd_v {sd= 0.001} 1.530
eval ($pd_v=$pd_x* 16000.0) BOND CMAL CMAF $pd_v {sd= 0.001} 1.530
eval ($pd_v=$pd_x* 18000.0) BOND CMAL NMAI $pd_v {sd= 0.001} 1.468
eval ($pd_v=$pd_x* 16000.0) BOND CMAL CMAJ $pd_v {sd= 0.001} 1.516
eval ($pd_v=$pd_x* 12000.0) BOND NMAI OMAB $pd_v {sd= 0.001} 1.318
eval ($pd_v=$pd_x* 18000.0) BOND NMAI CMAK $pd_v {sd= 0.001} 1.463
eval ($pd_v=$pd_x* 15000.0) BOND OMAB MMAA $pd_v {sd= 0.001} 1.000
eval ($pd_v=$pd_x* 16000.0) BOND CMAK CMAC $pd_v {sd= 0.001} 1.530
eval ($pd_v=$pd_x* 16000.0) BOND CMAK CMAD $pd_v {sd= 0.001} 1.530
eval ($pd_v=$pd_x* 16000.0) BOND CMAK CMAG $pd_v {sd= 0.001} 1.506
eval ($pd_v=$pd_x* 16000.0) BOND CMAG CMAJ $pd_v {sd= 0.001} 1.338
eval ($pd_v=$pd_x* 16000.0) BOND CMAJ CMAH $pd_v {sd= 0.001} 1.530
eval ($pd_v=$pd_x* 18000.0) BOND CMAH SMAA $pd_v {sd= 0.001} 1.830
eval ($pd_v=$pd_x* 15000.0) BOND SMAA SH1E $pd_v {sd= 0.001} 2.030
eval ($pd_v=$pd_x* 15000.0) BOND CMAG MMA $pd_v {sd= 0.001} 1.000
BOND CMAH MMA 1000.000 {sd= 0.001} 1.080
BOND CMAC MMA 1000.000 {sd= 0.001} 1.080
BOND CMAD MMA 1000.000 {sd= 0.001} 1.080
BOND CMAE MMA 1000.000 {sd= 0.001} 1.080
BOND CMAF MMA 1000.000 {sd= 0.001} 1.080
! BOND CH2E SH1E 1000.000 {sd= 0.001} 1.808
eval ($pd_v=$pd_x* 880.0) ANGLE CMAE CMAL CMAF $pd_v {sd= 0.031} 109.130
eval ($pd_v=$pd_x* 880.0) ANGLE CMAE CMAL NMAI $pd_v {sd= 0.031} 108.044
eval ($pd_v=$pd_x* 760.0) ANGLE CMAE CMAL CMAJ $pd_v {sd= 0.031} 111.354
eval ($pd_v=$pd_x* 880.0) ANGLE CMAF CMAL NMAI $pd_v {sd= 0.031} 113.418
eval ($pd_v=$pd_x* 760.0) ANGLE CMAF CMAL CMAJ $pd_v {sd= 0.031} 109.251
eval ($pd_v=$pd_x* 880.0) ANGLE NMAI CMAL CMAJ $pd_v {sd= 0.031} 105.635
eval ($pd_v=$pd_x* 720.0) ANGLE CMAL NMAI OMAB $pd_v {sd= 0.031} 115.650
eval ($pd_v=$pd_x* 880.0) ANGLE CMAL NMAI CMAK $pd_v {sd= 0.031} 104.000
eval ($pd_v=$pd_x* 720.0) ANGLE OMAB NMAI CMAK $pd_v {sd= 0.031} 106.714
eval ($pd_v=$pd_x* 760.0) ANGLE NMAI OMAB MMAA $pd_v {sd= 0.031} 109.500
eval ($pd_v=$pd_x* 880.0) ANGLE NMAI CMAK CMAC $pd_v {sd= 0.031} 113.839
eval ($pd_v=$pd_x* 880.0) ANGLE NMAI CMAK CMAD $pd_v {sd= 0.031} 109.317
eval ($pd_v=$pd_x* 880.0) ANGLE NMAI CMAK CMAG $pd_v {sd= 0.031} 105.283
eval ($pd_v=$pd_x* 880.0) ANGLE CMAC CMAK CMAD $pd_v {sd= 0.031} 109.700
eval ($pd_v=$pd_x* 760.0) ANGLE CMAC CMAK CMAG $pd_v {sd= 0.031} 108.346
eval ($pd_v=$pd_x* 760.0) ANGLE CMAD CMAK CMAG $pd_v {sd= 0.031} 110.253
eval ($pd_v=$pd_x* 800.0) ANGLE CMAK CMAG CMAJ $pd_v {sd= 0.031} 110.468
eval ($pd_v=$pd_x* 800.0) ANGLE CMAL CMAJ CMAG $pd_v {sd= 0.031} 108.765
eval ($pd_v=$pd_x* 800.0) ANGLE CMAL CMAJ CMAH $pd_v {sd= 0.031} 120.000
eval ($pd_v=$pd_x* 800.0) ANGLE CMAG CMAJ CMAH $pd_v {sd= 0.031} 126.000
eval ($pd_v=$pd_x* 880.0) ANGLE CMAJ CMAH SMAA $pd_v {sd= 0.031} 111.000
eval ($pd_v=$pd_x* 760.0) ANGLE CMAH SMAA SH1E $pd_v {sd= 0.031} 103.8
eval ($pd_v=$pd_x* 760.0) ANGLE SMAA SH1E CH2E $pd_v {sd= 0.031} 103.8 !yizhou
eval ($pd_v=$pd_x* 800.0) ANGLE MMA CMAG CMAK $pd_v {sd= 0.031} 126.8575 !yizhou
eval ($pd_v=$pd_x* 800.0) ANGLE MMA CMAG CMAJ $pd_v {sd= 0.031} 122.675 !yizhou
! ANGLE SM1E CH2E SM1E 0.0 {sd= 0.031} 0.000
ANGLE SMAA SH1E SMAA 0.0 {sd= 0.031} 110.000
ANGLe MMA CMAH MMA 500.00 {sd= 0.031} 109.4074
ANGLe MMA CMAC MMA 500.00 {sd= 0.031} 109.4703
ANGLe MMA CMAD MMA 500.00 {sd= 0.031} 109.4703
ANGLe MMA CMAE MMA 500.00 {sd= 0.031} 109.4703
ANGLe MMA CMAF MMA 500.00 {sd= 0.031} 109.4703
ANGLe CMAJ CMAH MMA 500.00 {sd= 0.031} 108.7236
ANGLe MMA CMAH SMAA 500.00 {sd= 0.031} 107.9228
ANGLe CMAL CMAE MMA 500.00 {sd= 0.031} 109.4726
ANGLe CMAL CMAF MMA 500.00 {sd= 0.031} 109.4726
ANGLe CMAK CMAC MMA 500.00 {sd= 0.031} 109.4726
ANGLe CMAK CMAD MMA 500.00 {sd= 0.031} 109.4726
! ANGLe HA CH2E SM1E 500.00 {sd= 0.031} 107.9185
! ANGLe CH1E CH2E SM1E 500.00 {sd= 0.031} 114.3558
eval ($pd_v=$pd_x* 800.0) IMPR CMAJ CMAL CMAG CMAH $pd_v 0 0.000
eval ($pd_v=$pd_x* 400.0) IMPR CMAL CMAE CMAF NMAI $pd_v 0 35.264
eval ($pd_v=$pd_x* 400.0) IMPR NMAI CMAL OMAB CMAK $pd_v {sd= 2.0} 0 -35.264
eval ($pd_v=$pd_x* 400.0) IMPR CMAK NMAI CMAC CMAD $pd_v {sd= 0.031} 0 35.264
IMPR MMA MMA SMAA CMAJ 500.00 {sd= 0.031} 0 -70.7825
IMPR MMA MMA CMAL MMA 500.00 {sd= 0.031} 0 -66.5692
IMPR MMA MMA CMAK MMA 500.00 {sd= 0.031} 0 -66.5692
! IMPRoper HA HA CH1E SM1E 500.00 {sd= 0.031} 0 -72.0234
eval ($pd_v=$pd_x* 0) DIHE CMAE CMAL NMAI CMAK $pd_v 3 0.000
eval ($pd_v=$pd_x* 0) DIHE CMAE CMAL CMAJ CMAH $pd_v 6 0.000
!eval ($pd_v=$pd_x* 0.9) DIHE CMAL NMAI OMAB MMAA $pd_v 3 0.000
eval ($pd_v=$pd_x* 8.1) DIHE CMAG CMAK NMAI CMAL $pd_v 3 0.000
eval ($pd_v=$pd_x* 0.1) DIHE NMAI CMAK CMAG CMAJ $pd_v 6 0.000
eval ($pd_v=$pd_x* 19.6) DIHE CMAK CMAG CMAJ CMAH $pd_v 2 180.000
eval ($pd_v=$pd_x* 0) DIHE CMAL CMAJ CMAH SMAA $pd_v 6 0.000
!eval ($pd_v=$pd_x* 4.9) DIHE SM1E SMAA CMAH CMAJ $pd_v 3 0.000
eval ($pd_v=$pd_x* 19.6) DIHE MMA CMAG CMAJ CMAL $pd_v 2 180.000 !yizhou
eval ($pd_v=$pd_x* 19.6) DIHE OMAB NMAI CMAL CMAJ $pd_v {sd= 0.031} 2 -146.7663 !yizhou
! DIHEdral NH1 CH1E CH2E SM1E 0.00 {sd= 0.031} 3 0.0000
!NBONds
! TOLERANCE=0.5 NBXMOD=5 WMIN=1.5
! REPEL=1.0 REXPONENT=4 IREXPONENT=1 RCONST=16.0
! CTONNB=5.5 CTOFNB=6.0 CUTNB=7.0
!END
NONBONDED CMAE 0.10000 3.29633 0.10000 3.02906
NONBONDED CMAL 0.10000 3.29633 0.10000 3.02906
NONBONDED CMAF 0.10000 3.29633 0.10000 3.02906
NONBONDED NMAI 0.10000 2.67270 0.10000 2.40543
NONBONDED OMAB 0.10000 2.58361 0.10000 2.31634
NONBONDED MMAA 0.10000 1.42544 0.10000 1.15817
NONBONDED CMAK 0.10000 3.29633 0.10000 3.02906
NONBONDED CMAC 0.10000 3.29633 0.10000 3.02906
NONBONDED CMAD 0.10000 3.29633 0.10000 3.02906
NONBONDED CMAG 0.10000 3.29633 0.10000 3.02906
NONBONDED CMAJ 0.10000 3.29633 0.10000 3.02906
NONBONDED CMAH 0.10000 3.29633 0.10000 3.02906
NONBONDED SMAA 0.10000 3.20724 0.10000 2.93997
NONBONDED MMA 0.10000 1.42544 0.10000 1.15817
!NONBONDED SM1E 0.10000 3.20724 0.10000 2.93997
Invoking pseudo-residues
In the protein sequence file, use “CYSM” to represent a cysteine residue conjugated to a MTSL molecule. If MTSL chain mobility is considered for ensemble averaging, use “C2M” or “C3M”, which allows averaging for 2 or 3 MTSL conformers.
Generate psf/mtf file and an extended pdb structure from the primary sequence, and examine the pseudo-residue structure by your favorite PDB viewer.
Ensemble averaging of PRE
Basics
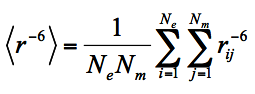 (1)
(1)
Where Ne is the number of ensemble states, i.e. the number of conformers the protein has, Nm is the number of MTSL conformers used to represent chain mobility, e.g. Nm =3 if C3M is used, rij is the distance between the unpaired electron (approximated by MTSL oxygen OAB/OBB/OCB) and a proton for the i-th protein conformer and the j-th MTSL conformer.
To relate <r-6> to PRE, use Solomon-Bloembergen (SB) equation for delta R2:
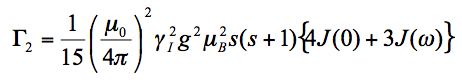 (2)
(2)
Where,
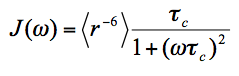 (3)
(3)
Range of validity: Eq(3) is a good approximation when the amplitude of internal motions, including the motion among the protein and MTSL conformers, is small or when the time-scale of these motions is significantly longer than that of the global tumbling. For motions of both large amplitude and short time-scale, a more accurate description is by the SBMF equation [5]. It should be noted that the error from Eq(3) in the latter case is in the relaxation space, i.e., in <r-6>. The propagated error in the distance space r, which is of more interest to us for structure purpose, is usually quite small.
Implementation in Xplor-nih
Dealing with averaging
Xplor-nih-python provides a nice interface for using ensemble averaged potential energy.
To create such an instance,
from ensembleSimulation import EnsembleSimulation
esim = EnsembleSimulation("ensemble",ensembleSize)
ensembleSize is an integer number which specifies the number of alternative protein conformers, i.e., Ne in Eq (1).
To add an energy term to the potential list, such as PRE, do the following
(Read the inline comments on certain commands):
potList = PotList() !initiate the potential list for esim. This needs to be done only once
import prePot
pre1=prePot.PREPot("PRE_CT_1",open("pre_CT_1.tbl").read(),"normal")
pre2=prePot.PREPot("PRE_CT_2",open("pre_CT_2.tbl").read(),"normal")
pre3=prePot.PREPot("PRE_NT_1",open("pre_NT_1.tbl").read(),"normal")
pre4=prePot.PREPot("PRE_NT_2",open("pre_NT_2.tbl").read(),"normal")
! There are 4 pre input files. Read them into 4 pre pot terms, pre1, pre2, pre3, and pre4.
! You can combine them into one, but keeping separate is easier to manage.
allpre = (pre1,pre2,pre3,pre4)
tauc=30 ! tauc is 30ns. Tauc can be measured or estimated based on protein size.
for pre in allpre: !Define equation and parameters for PRE back-calcualtion
pre.setEquType("sb") !Use Solomon-Bloembergen equation
pre.setAveType("r-6") ! Averaging type for ambiguous PRE assignment
pre.setSclType("obsig")
pre.setRlxType("r2dd")
pre.setGammaI(26.752196)
pre.setSqn(0.5)
pre.setGfac(2.0)
pre.setTcType("fix")
pre.setTauC(tauc)
print " setting for ", pre.instanceName()
potList.add(pre) ! Add pre into potlist for ensemble averaging.
pass
Note that so far we only specified averaging of protein conformers, but we haven’t done so for MTSL conformers. To do this, we can use ambiguous assignment in the pre input file, such as “pre_CT_1.tbl”
assign (resid 149 and name HN) (resid 117 and (name OAB or name OBB or name OCB)) 106.2 6.1
Where, resid 117 is a C3M residue. The 106.2 ± 6.1 s-1 PRE on HN of resid 149 is <r-6> averaged by the 3 MTSL conformers with electron position represented by OAB, OBB, and OCB respectively. This averaging type is specified by “pre.setAveType("r-6")” in the pre setup loop.
Remove van der waals interaction among MTSL conformers
Note that the MTSL conformers are used to simulate motions, so they must not interfere with each other. Meanwhile, multiple MTSL labels are not simultaneously on the protein, therefore they shouldn’t interfere either. However, MTSL clash with native residues on the protein should be avoided. To express this in xplor language:
command(“””
vector identity ( store1 ) (chemical MM* or chemical CM* or chemical OM* or chemical NM* or chemical SM*)
vector identity ( store2 ) (known and not (store1 or (resname ANI)))
constraints
interaction (store1) (store1) weights * 1 vdw 0 end
interaction (store2) (known and not (resname ANI)) weights * 1 angl %f impr %f
end
“””)
Semi-Rigid-body dynamics
PRE is frequently used to dock two proteins of known structures. In this case, both proteins can be treated as rigid bodies during simulated annealing while the MTSL chains are variable. To realize this:
command("""
vector identity (store8) (name N or name HN or name CA or name HA or name C or name O)
""")
dyn.group( select('resid 17:178 and ((store8) or not (resid 176 or resid 117 or resid 83 or resid 55 or resid 59)) '))
In the example above, there are 5 C3M residues, 176, 117, 83, 55, and 59. Their side-chains are mobile. All other residues are grouped and therefore rigid.
To ensemble or not to ensemble
Some potential terms are not intended for ensemble averaging. Most of these are generic Xplor potentials, such as bond and angle energies. To signify this, use the “AvePot” command before adding them to the potential list.
potList.append( AvePot(XplorPot,"BOND") )
As a second example, NOEs may be treated as averaged between different protein conformers, or simply treated as related to each member individually, depending on how you want to treat them. For the former case,
enoe = create_NOEPot('enoe','noe.tbl')
potList.append( enoe )
For the latter case,
potList.append( AvePot(XplorPot,"NOE") )
References
2. Schwieters, C.D., Kuszewski, J.J., and Clore, G.M. (2006). Using Xplor-NIH for NMR molecular structure determination. Prog. NMR Spect. 48, 47-62.