MolProbity Server: Difference between revisions
No edit summary |
No edit summary |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== '''Introduction''' == | == '''Introduction''' == | ||
The MolProbity server, developed by the Richardson group | The MolProbity server, developed by the Richardson group [1,2], is a site for the evaluation of single X-ray structures and NMR structure ensembles. The server performs a number of validations including all-residue Ramachandran analysis, rotamer analysis, and all-atom clash analysis. Like the PSVS server, the MolProbity server is a valuable structure validation tool in the final stages of structure refinement. Structures and various metrics are visualized in the KiNG tool, and all analyses and “kinemages” can be downloaded. The server can be accessed at: [http://molprobity.biochem.duke.edu/ http://molprobity.biochem.duke.edu/]. | ||
== '''Using MolProbity server''' == | == '''Using MolProbity server''' == | ||
=== Uploading a PDB === | === Uploading a PDB === | ||
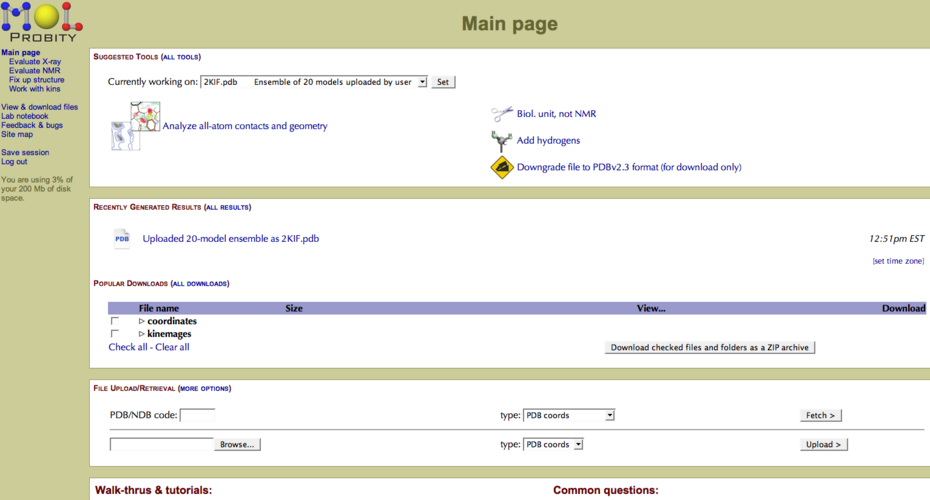
PDB files are quickly uploaded, and in the case of NMR ensembles, divided into individual PDB files. One next analyzes all-atom contacts and geometry (Figure 1). There is also an “add hydrogens” tool for X-ray structures, which is useful for comparison of NMR/X-ray structure pairs. | PDB files are quickly uploaded, and in the case of NMR ensembles, divided into individual PDB files. One next analyzes all-atom contacts and geometry (Figure 1). There is also an “add hydrogens” tool for X-ray structures, which is useful for comparison of NMR/X-ray structure pairs. | ||
===== Figure 1: File upload and analysis options in MolProbity ===== | ===== Figure 1: File upload and analysis options in MolProbity ===== | ||
[[Image: | [[Image:MolProbity upload.png|1000x500px]] | ||
=== Structure Analysis in KiNG === | === Structure Analysis in KiNG === | ||
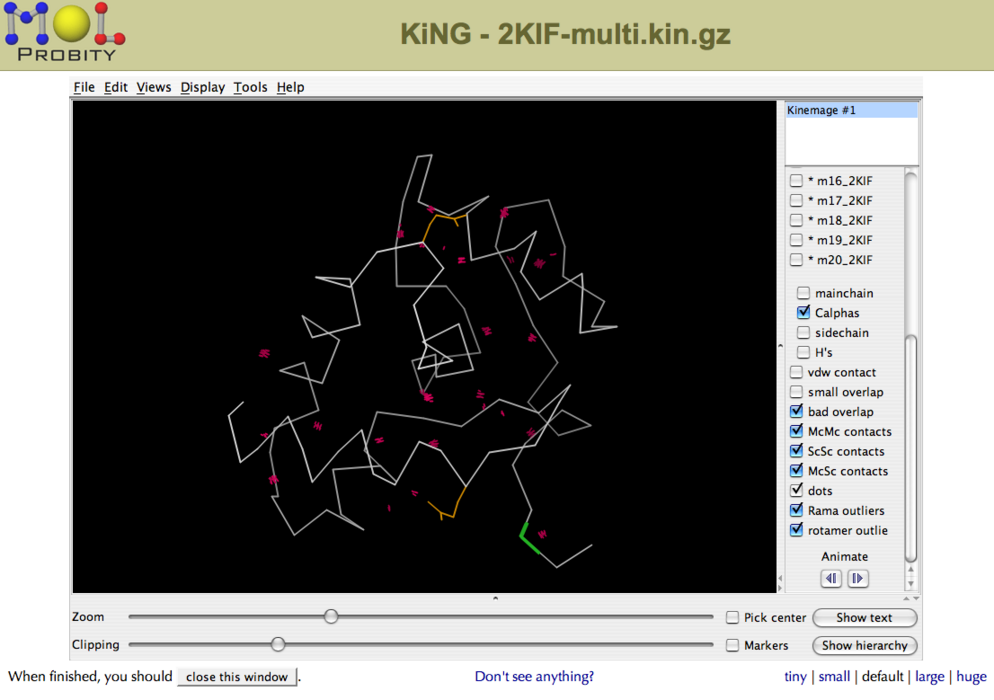
Structures can be analyzed using the interactive 3D software package KiNG (Figure 2). The kinemage files feature various structure validation metrics mapped onto the structures(s), including close contacts (pink dots), Ramachandran outliers (green lines) and bad rotamers (yellow side chains). | Structures can be analyzed using the interactive 3D software package KiNG (Figure 2). The kinemage files feature various structure validation metrics mapped onto the structures(s), including close contacts (pink dots), Ramachandran outliers (green lines) and bad rotamers (yellow side chains). | ||
===== Figure 2: Interactive structure analysis in KiNG<br> ===== | ===== Figure 2: Interactive structure analysis in KiNG<br> ===== | ||
[[Image: | [[Image:MolProbity KiNG.png|1000x700px]] | ||
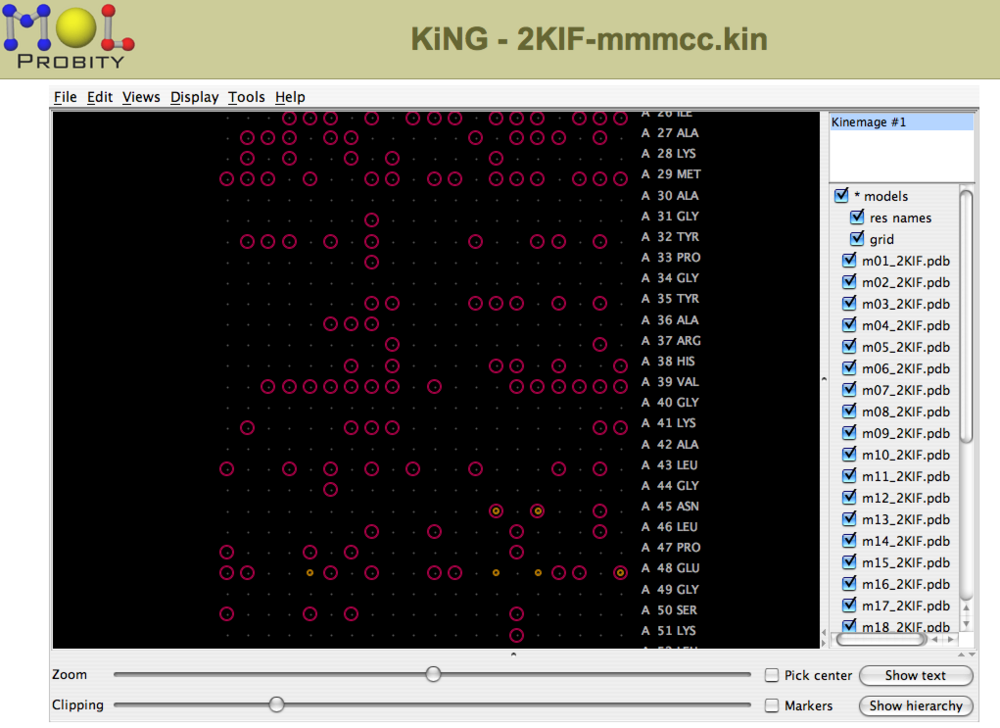
For NMR structure ensembles, violations can be viewed for each residue in each structure in a multichart format (Figure 3). | For NMR structure ensembles, violations can be viewed for each residue in each structure in a multichart format (Figure 3). | ||
===== Figure 3: Multi-chart analysis of outliers and clashes<br> ===== | ===== Figure 3: Multi-chart analysis of outliers and clashes<br> ===== | ||
[[Image: | [[Image:MolProbity multichart.png|1000x750px]] | ||
A gzipped kinemage file can be downloaded for archiving and future analysis. The stand-alone version of the KiNG program can be downloaded at: [http://kinemage.biochem.duke.edu/ http://kinemage.biochem.duke.edu/].<br> | A gzipped kinemage file can be downloaded for archiving and future analysis. The stand-alone version of the KiNG program can be downloaded at: [http://kinemage.biochem.duke.edu/ http://kinemage.biochem.duke.edu/].<br> | ||
=== Ramachandran Analysis === | === Ramachandran Analysis === | ||
The MolProbity output also includes a complete Ramachandran analysis of all residues in all models, which can be downloaded as a pdf file. Here is an example Ramachandran output file: [[ | The MolProbity output also includes a complete Ramachandran analysis of all residues in all models, which can be downloaded as a pdf file. Here is an example Ramachandran output file: [[Media:2KIF-rama.pdf|2KIF-rama.pdf]].<br> | ||
== '''References''' == | == '''References''' == | ||
1. Davis, I.W., Leaver-Fay, A., Chen, V.B., Block, J.N., Kapral, G.J., Wang, X., Murray, L.W., Arendall III, W.B., Snoeyink, J., Richardson, J.S. and Richardson, D.C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. ''Nucleic Acids Res. 35'', W375-W383.<br>2. Lovell, S.C., Davis, I.W., Arendall III, W.B., de Bakker, P.I.W., Word, J.M., Prisant, M.G., Richardson, J.S. and Richardson, D.C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. ''Proteins 50'', 437-450. | [http://www.ncbi.nlm.nih.gov/pubmed/17452350?itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum&ordinalpos=2 1. Davis, I.W., Leaver-Fay, A., Chen, V.B., Block, J.N., Kapral, G.J., Wang, X., Murray, L.W., Arendall III, W.B., Snoeyink, J., Richardson, J.S. and Richardson, D.C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. ''Nucleic Acids Res. 35'', W375-W383.]<br>[http://www3.interscience.wiley.com/journal/102523245/abstract?CRETRY=1&SRETRY=0 2. Lovell, S.C., Davis, I.W., Arendall III, W.B., de Bakker, P.I.W., Word, J.M., Prisant, M.G., Richardson, J.S. and Richardson, D.C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. ''Proteins 50'', 437-450.] | ||
<br> | |||
<br><br> | |||
Latest revision as of 17:00, 5 January 2010
Introduction
The MolProbity server, developed by the Richardson group [1,2], is a site for the evaluation of single X-ray structures and NMR structure ensembles. The server performs a number of validations including all-residue Ramachandran analysis, rotamer analysis, and all-atom clash analysis. Like the PSVS server, the MolProbity server is a valuable structure validation tool in the final stages of structure refinement. Structures and various metrics are visualized in the KiNG tool, and all analyses and “kinemages” can be downloaded. The server can be accessed at: http://molprobity.biochem.duke.edu/.
Using MolProbity server
Uploading a PDB
PDB files are quickly uploaded, and in the case of NMR ensembles, divided into individual PDB files. One next analyzes all-atom contacts and geometry (Figure 1). There is also an “add hydrogens” tool for X-ray structures, which is useful for comparison of NMR/X-ray structure pairs.
Figure 1: File upload and analysis options in MolProbity
Structure Analysis in KiNG
Structures can be analyzed using the interactive 3D software package KiNG (Figure 2). The kinemage files feature various structure validation metrics mapped onto the structures(s), including close contacts (pink dots), Ramachandran outliers (green lines) and bad rotamers (yellow side chains).
Figure 2: Interactive structure analysis in KiNG
For NMR structure ensembles, violations can be viewed for each residue in each structure in a multichart format (Figure 3).
Figure 3: Multi-chart analysis of outliers and clashes
A gzipped kinemage file can be downloaded for archiving and future analysis. The stand-alone version of the KiNG program can be downloaded at: http://kinemage.biochem.duke.edu/.
Ramachandran Analysis
The MolProbity output also includes a complete Ramachandran analysis of all residues in all models, which can be downloaded as a pdf file. Here is an example Ramachandran output file: 2KIF-rama.pdf.
References
1. Davis, I.W., Leaver-Fay, A., Chen, V.B., Block, J.N., Kapral, G.J., Wang, X., Murray, L.W., Arendall III, W.B., Snoeyink, J., Richardson, J.S. and Richardson, D.C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375-W383.
2. Lovell, S.C., Davis, I.W., Arendall III, W.B., de Bakker, P.I.W., Word, J.M., Prisant, M.G., Richardson, J.S. and Richardson, D.C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50, 437-450.