FOUND: Difference between revisions
No edit summary |
No edit summary |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== '''Introduction''' == | |||
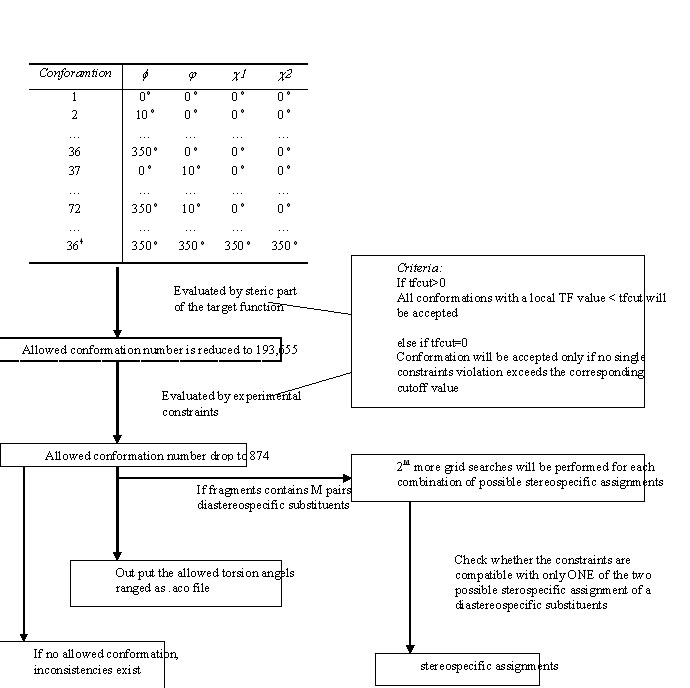
'''FOUND''' performs systematic local conformation analysis by exhaustive grid searches for arbitrary contiguous fragments of proteins and nucleic acids. It uses torsion angles as the only degrees of freedom to identify all conformations that fulfill the steric and NMR-derived conformational restraints within a contiguous molecular fragment, as defined either by limits on the maximal restraint violations or by the fragment-based target function value. | '''FOUND''' performs systematic local conformation analysis by exhaustive grid searches for arbitrary contiguous fragments of proteins and nucleic acids. It uses torsion angles as the only degrees of freedom to identify all conformations that fulfill the steric and NMR-derived conformational restraints within a contiguous molecular fragment, as defined either by limits on the maximal restraint violations or by the fragment-based target function value. | ||
| Line 16: | Line 13: | ||
angles=side-chain angles (default: CHI1) | angles=side-chain angles (default: CHI1) | ||
tfcut=fmax (default: 0.0) | tfcut=fmax (default: 0.0) | ||
continue</pre> | continue</pre> | ||
<br> Performs for all amino acid residues in the given range grid searches comprising the backbone dihedral angles φ, ψ, χ1 and the given side-chain angles. To specify more than one side-chain angle, the names must be given, separated by blanks and enclosed in double quotes. If the cutoff value for the local, fragment-based target function, <tt>fmax</tt>, is positive, then all conformations with a local target function value below <tt>fmax</tt> will be considered as allowed. Otherwise, a conformation will be allowed if no single restraint violation exceeds the corresponding cutoff value defined by the variables <tt>soft_upl</tt>, <tt>soft_lol</tt>, etc. Unless the <tt>continue</tt> option is set, the allowed ranges of dihedral angles will be initialized to allow all possible angle values before the grid searches are started. | <br> Performs for all amino acid residues in the given range grid searches comprising the backbone dihedral angles φ, ψ, χ1 and the given side-chain angles. To specify more than one side-chain angle, the names must be given, separated by blanks and enclosed in double quotes. If the cutoff value for the local, fragment-based target function, <tt>fmax</tt>, is positive, then all conformations with a local target function value below <tt>fmax</tt> will be considered as allowed. Otherwise, a conformation will be allowed if no single restraint violation exceeds the corresponding cutoff value defined by the variables <tt>soft_upl</tt>, <tt>soft_lol</tt>, etc. Unless the <tt>continue</tt> option is set, the allowed ranges of dihedral angles will be initialized to allow all possible angle values before the grid searches are started. | ||
FOUND is implemented as a CYANA and DYANA macro succeeding the standalone HABAS program. Therefore, the FOUND macros are called <tt>habas.cya</tt> or <tt>habas.dya</tt>. For more information type <tt>help habas</tt> in CYANA or see the reference below. | FOUND is implemented as a CYANA and DYANA macro succeeding the standalone HABAS program. Therefore, the FOUND macros are called <tt>habas.cya</tt> or <tt>habas.dya</tt>. For more information type <tt>help habas</tt> in CYANA or see the reference below. | ||
References:<br> FOUND - [http://www.springerlink.com/content/t318k12296032kk5/fulltext.pdf Guntert P. ''J. Biomol. NMR'' 12 (4): 543-548 NOV 1998]<br> HABAS - [http://pubs.acs.org/cgi-bin/archive.cgi/jacsat/1989/111/i11/pdf/ja00193a036.pdf Guntert P. ''J. Am. Chem. Soc.'' 1989, 111, 3997-4004] | References:<br> FOUND - [http://www.springerlink.com/content/t318k12296032kk5/fulltext.pdf Guntert P. ''J. Biomol. NMR'' 12 (4): 543-548 NOV 1998]<br> HABAS - [http://pubs.acs.org/cgi-bin/archive.cgi/jacsat/1989/111/i11/pdf/ja00193a036.pdf Guntert P. ''J. Am. Chem. Soc.'' 1989, 111, 3997-4004] | ||
<br> | |||
== '''How FOUND (HABAS) Works'''<br> == | |||
[[Image:Found3.jpg]]<br> | |||
<br> | |||
== '''Preparing to run FOUND (HABAS)''' == | |||
If you are using completely unassigned peaklists and you don't have any J-coupling data, then you don't need to run found. Otherwise, do the following: | |||
< | #Create a working subdirectory (for example, <tt>structure/cyana21/found</tt>). | ||
protocol:=found.out | #If you have ct-[<sup>13</sup>C,<sup>1</sup>H]-HSQC spectra for the fractionally <sup>13</sup>C-labeled sample, determine the stereospecific assignments for Val and Leu methyl groups and swap spin labels (in the XEASY atom list or CARA repository) where necessary, as described in [[NESG:SSAFromFractional13CLabeledSample|Stereospecific assignments from fractionally 13C-labeled sample]]. Create a file <tt>stereo.cya</tt> to declare the residues, for which stereospecific assignments were established. | ||
#Copy the latest sequence (<tt>XXXX.seq</tt>), atomlist (<tt>XXXX.prot</tt>) and peaklist files (<tt>n.peaks</tt>, <tt>ali.peaks</tt> and <tt>aro.peaks</tt>) into the working directory. Modify the sequence file if you have non-standard residues, such as cis-proline (use <tt>cPRO</tt>). | |||
#Create an init.cya file as described in "[[NESG:CYANAInitFile|Creating an init.cya file for CYANA 2.1]]" or copy a previously used file. Set an appropriate RMSD calculation range. | |||
#Create a <tt>GridSearch.cya</tt> file or download a template [[Media:GridSearch.cya|GridSearch.cya]] (see below). | |||
#Copy the TALOS dihedral angel constraints file (<tt>talos.aco</tt>) from the previous step. Keep only those constraints, which correspond to residues in the secondary structure elements according to CSI, excluding one residue at the edge of each element. You can comment out the remaining lines with a hash character =#= using any text editor. | |||
#Produce initial calibration as described in NOECalibrationCYANA#Finding_Initial_NOE_Calibration. | |||
#Download the [[Media:CALC_found.cya|CALC.cya]] file to run a test structure calculation using the output of FOUND (see below). | |||
Here is the template <tt>GridSearch.cya</tt> script adapted from <tt>~/demos/details/GridSearch.cya</tt> in CYANA 2.1 installation directory. The options for scalar couplings have been commented out since they aren't routinely used. | |||
<pre>protocol:=found.out | |||
read seq $name.seq | read seq $name.seq | ||
read upl short.upl # read noe upper distance limits | read upl short.upl # read noe upper distance limits | ||
| Line 58: | Line 57: | ||
savestereo stereofound.cya # write macro with stereospecific assignments | savestereo stereofound.cya # write macro with stereospecific assignments | ||
protocol:= | protocol:= | ||
</ | </pre> | ||
<br> It is recommended to apply [[NESG:SSAFromFractional13CLabeledSample|stereospecific assignments of Leu and Val isopropyl groups]] (if available) when running HABAS. This may help HABAS derive additional stereospecific assignments for HB2/3 spins of Leu. To take advantage of stereospecific assignments create <tt>swap.cya</tt> and <tt>stereo.cya</tt> macros as described in the topic about [[NESG:ApplyingSSA|applying stereospecific assignments]] and use them to generate a swapped proton list and a swapped UPL file <tt>short.upl</tt> | |||
'''N.B.''' The <tt>GridSearch.cya</tt> script is not the same as the <tt>grid search</tt> command! | |||
<br> | |||
== '''Verifying Local NOE Network''' == | |||
# In CYANA 2.1 run the <tt>GridSearch.cya</tt> script. | #In CYANA 2.1 run the <tt>GridSearch.cya</tt> script. | ||
# In CYANA 2.1 run the <tt>CALC.cya</tt> script. This may take several minutes on a workstation. | #In CYANA 2.1 run the <tt>CALC.cya</tt> script. This may take several minutes on a workstation. | ||
# Go residue-by-residue through the FOUND output. If inconsistent data was found, double-check the NOE peaks for this residue in XEASY or CARA. UPL violations reported in the <tt>XXXX.ovw</tt> file usually help to pinpoint the questionable peaks | #Go residue-by-residue through the FOUND output. If inconsistent data was found, double-check the NOE peaks for this residue in XEASY or CARA. UPL violations reported in the <tt>XXXX.ovw</tt> file usually help to pinpoint the questionable peaks. | ||
**Usually ''inconsistent data'' reports are caused by peaks with improper volumes or incorrectly assigned peaks. Check assignments and integration in XEASY or CARA. | |||
* | **Too many ''inconsistent data'' reports and violations for apparently valid peaks indicate that calibration that is too tight. Look at the violations in the <tt>XXXX.ovw</tt> file to determine which peaklist requires calibration adjustment. Change calibration constants and re-run the calibration. | ||
* <tt> | |||
#Repeat steps 1 and 3 until no inconsistent data was found by FOUND and the average target function is < 0.5 . | |||
FOUND will produce the following files: | |||
[[Image:Found6.jpg]] | *<tt>stereofound.cya</tt> - CYANA script, which delcares stereospecific assignments. It is a combination of input stereospecific assignments and those generated by FOUND. | ||
*<tt>swapped.upl</tt> - a modification of the input UPL file, in which spin labels have been swapped according to the newly derived stereospecific assignments. This file itself is not used in subsequent automated structure calculation and NOE assignment, because of incompatible NOE calibration protocols. | |||
*<tt>gridsearch.aco</tt> - is a combination of input TALOS constraints and constraints generated with the grid search method. | |||
<br> | |||
== '''Stereospecific Assignments from FOUND (HABAS)''' == | |||
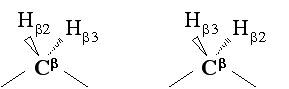
Stereo-specific assignment can be obtain from checking FOUND module without any preliminary structure. FOUND module performing separate grid searches for both possible stereospecific assignments for diastereotopic protons or isopropyl groups as show in the figure below:<br> | |||
[[Image:Found6.jpg]] | |||
where the allowed conformation numbers for the two possible stereospecific assignments are N1 and N2, separately | where the allowed conformation numbers for the two possible stereospecific assignments are N1 and N2, separately | ||
| Line 106: | Line 111: | ||
45 MET HB2 HB3 assigned | 45 MET HB2 HB3 assigned | ||
45 MET HG2 HG3 | 45 MET HG2 HG3 | ||
</pre> | </pre> | ||
See the data upstream from the summary to find whether atom pair were assigned "swapped" or "as input". Verify that peaks assigned to the reported spins are integrated correctly. Avoid accepting assignments with very few allowed conformations (such as Met45 below), because they are indicative of too tight UPLs. | See the data upstream from the summary to find whether atom pair were assigned "swapped" or "as input". Verify that peaks assigned to the reported spins are integrated correctly. Avoid accepting assignments with very few allowed conformations (such as Met45 below), because they are indicative of too tight UPLs. | ||
<pre> ARG 33: | <pre> ARG 33: | ||
| Line 115: | Line 120: | ||
HB2/HB3 MET 45: 35/0 conformations, assigned as input. | HB2/HB3 MET 45: 35/0 conformations, assigned as input. | ||
HG2/HG3 MET 45: 14/21 conformations, unassigned. | HG2/HG3 MET 45: 14/21 conformations, unassigned. | ||
</pre> | </pre> | ||
Delete or comment out those lines in <tt>stereofound.cya</tt>, which you think correspond to incorrectly derived stereospecific assignments. | Delete or comment out those lines in <tt>stereofound.cya</tt>, which you think correspond to incorrectly derived stereospecific assignments. | ||
In subsequent steps it is recommended to swap the labels in the original source containing chemical shift assignments: the atom list if using XEASY, or CARA repository if using CARA. | In subsequent steps it is recommended to swap the labels in the original source containing chemical shift assignments: the atom list if using XEASY, or CARA repository if using CARA. | ||
<br> | |||
<br> | |||
<br> | <br> | ||
*[[ | *[[Media:GridSearch.cya|GridSearch.cya]]: CYANA script to run FOUND module | ||
*[[ | *[[Media:CALC_found.cya|CALC.cya]]: CYANA script to run manual calculation with local constraints | ||
Latest revision as of 21:48, 6 January 2010
Introduction
FOUND performs systematic local conformation analysis by exhaustive grid searches for arbitrary contiguous fragments of proteins and nucleic acids. It uses torsion angles as the only degrees of freedom to identify all conformations that fulfill the steric and NMR-derived conformational restraints within a contiguous molecular fragment, as defined either by limits on the maximal restraint violations or by the fragment-based target function value.
FOUND does the following:
- Detects inconsistencies in the experimental.
- Derives stereospecific assignments of diastereotopic methylene protons or isopropyl groups.
- Converts coupling constants and UPLs into torsion angle restraints.
Parameters:
range=residue range (default: all amino acid residues)
angles=side-chain angles (default: CHI1)
tfcut=fmax (default: 0.0)
continue
Performs for all amino acid residues in the given range grid searches comprising the backbone dihedral angles φ, ψ, χ1 and the given side-chain angles. To specify more than one side-chain angle, the names must be given, separated by blanks and enclosed in double quotes. If the cutoff value for the local, fragment-based target function, fmax, is positive, then all conformations with a local target function value below fmax will be considered as allowed. Otherwise, a conformation will be allowed if no single restraint violation exceeds the corresponding cutoff value defined by the variables soft_upl, soft_lol, etc. Unless the continue option is set, the allowed ranges of dihedral angles will be initialized to allow all possible angle values before the grid searches are started.
FOUND is implemented as a CYANA and DYANA macro succeeding the standalone HABAS program. Therefore, the FOUND macros are called habas.cya or habas.dya. For more information type help habas in CYANA or see the reference below.
References:
FOUND - Guntert P. J. Biomol. NMR 12 (4): 543-548 NOV 1998
HABAS - Guntert P. J. Am. Chem. Soc. 1989, 111, 3997-4004
How FOUND (HABAS) Works
Preparing to run FOUND (HABAS)
If you are using completely unassigned peaklists and you don't have any J-coupling data, then you don't need to run found. Otherwise, do the following:
- Create a working subdirectory (for example, structure/cyana21/found).
- If you have ct-[13C,1H]-HSQC spectra for the fractionally 13C-labeled sample, determine the stereospecific assignments for Val and Leu methyl groups and swap spin labels (in the XEASY atom list or CARA repository) where necessary, as described in Stereospecific assignments from fractionally 13C-labeled sample. Create a file stereo.cya to declare the residues, for which stereospecific assignments were established.
- Copy the latest sequence (XXXX.seq), atomlist (XXXX.prot) and peaklist files (n.peaks, ali.peaks and aro.peaks) into the working directory. Modify the sequence file if you have non-standard residues, such as cis-proline (use cPRO).
- Create an init.cya file as described in "Creating an init.cya file for CYANA 2.1" or copy a previously used file. Set an appropriate RMSD calculation range.
- Create a GridSearch.cya file or download a template GridSearch.cya (see below).
- Copy the TALOS dihedral angel constraints file (talos.aco) from the previous step. Keep only those constraints, which correspond to residues in the secondary structure elements according to CSI, excluding one residue at the edge of each element. You can comment out the remaining lines with a hash character =#= using any text editor.
- Produce initial calibration as described in NOECalibrationCYANA#Finding_Initial_NOE_Calibration.
- Download the CALC.cya file to run a test structure calculation using the output of FOUND (see below).
Here is the template GridSearch.cya script adapted from ~/demos/details/GridSearch.cya in CYANA 2.1 installation directory. The options for scalar couplings have been commented out since they aren't routinely used.
protocol:=found.out read seq $name.seq read upl short.upl # read noe upper distance limits stereo # apply stereospecific assignments from stereo.cya #read cco demo.cco # read J-coupling constants #karplus # use standard Karplus curves (from karplus.cya) #habas angles="CHI1" tfcut=0.05 # perform phi/psi/chi1 grid search (HABAS) habas angles="CHI1 CHI2*" tfcut=0.05 # alternative phi/psi/chi1/chi2 grid search gridplot habas.ps atom stereo list write upl swapped.upl # write upper limits with swapped stereo pairs write aco gridsearch.aco # write upper limits with swapped stereo pairs #write cco swapped.cco # write couplings with swapped stereo pairs savestereo stereofound.cya # write macro with stereospecific assignments protocol:=
It is recommended to apply stereospecific assignments of Leu and Val isopropyl groups (if available) when running HABAS. This may help HABAS derive additional stereospecific assignments for HB2/3 spins of Leu. To take advantage of stereospecific assignments create swap.cya and stereo.cya macros as described in the topic about applying stereospecific assignments and use them to generate a swapped proton list and a swapped UPL file short.upl
N.B. The GridSearch.cya script is not the same as the grid search command!
Verifying Local NOE Network
- In CYANA 2.1 run the GridSearch.cya script.
- In CYANA 2.1 run the CALC.cya script. This may take several minutes on a workstation.
- Go residue-by-residue through the FOUND output. If inconsistent data was found, double-check the NOE peaks for this residue in XEASY or CARA. UPL violations reported in the XXXX.ovw file usually help to pinpoint the questionable peaks.
- Usually inconsistent data reports are caused by peaks with improper volumes or incorrectly assigned peaks. Check assignments and integration in XEASY or CARA.
- Too many inconsistent data reports and violations for apparently valid peaks indicate that calibration that is too tight. Look at the violations in the XXXX.ovw file to determine which peaklist requires calibration adjustment. Change calibration constants and re-run the calibration.
- Repeat steps 1 and 3 until no inconsistent data was found by FOUND and the average target function is < 0.5 .
FOUND will produce the following files:
- stereofound.cya - CYANA script, which delcares stereospecific assignments. It is a combination of input stereospecific assignments and those generated by FOUND.
- swapped.upl - a modification of the input UPL file, in which spin labels have been swapped according to the newly derived stereospecific assignments. This file itself is not used in subsequent automated structure calculation and NOE assignment, because of incompatible NOE calibration protocols.
- gridsearch.aco - is a combination of input TALOS constraints and constraints generated with the grid search method.
Stereospecific Assignments from FOUND (HABAS)
Stereo-specific assignment can be obtain from checking FOUND module without any preliminary structure. FOUND module performing separate grid searches for both possible stereospecific assignments for diastereotopic protons or isopropyl groups as show in the figure below:
where the allowed conformation numbers for the two possible stereospecific assignments are N1 and N2, separately
- if N1 >0, N2 >0, stereospecific assignment is ambiguous
- if N1 >0, N2 =0, assigned as input
- if N1 =0, N2 >0, assigned as reversed
- if N1 =0, N2 =0, both assignment possibilities are disallowed - inconsistent data.
After running FOUND module check by using CYANA macro "GridSeach.cya" check the output file (e.g. see figure below) for stereospecific assignments. This fragment shows that stereospecific assignments were derived for HB2/3 of Arg33 and Met45. Note that externally declared stereospecific assignments, such as QD1/2 of Leu34 and QG1/2 of Val38 in this example, are also listed.
33 ARG HB2 HB3 assigned
33 ARG HG2 HG3
33 ARG HD2 HD3
33 ARG HH11 HH12
33 ARG HH21 HH22
34 LEU HB2 HB3
34 LEU QD1 QD2 assigned
36 PHE HB2 HB3
37 PRO HB2 HB3
37 PRO HG2 HG3
37 PRO HD2 HD3
38 VAL QG1 QG2 assigned
45 MET HB2 HB3 assigned
45 MET HG2 HG3
See the data upstream from the summary to find whether atom pair were assigned "swapped" or "as input". Verify that peaks assigned to the reported spins are integrated correctly. Avoid accepting assignments with very few allowed conformations (such as Met45 below), because they are indicative of too tight UPLs.
ARG 33:
HB2/HB3 ARG 33: 0/1465 conformations, assigned reversed, swapped.
HG2/HG3 ARG 33: 698/767 conformations, unassigned.
...
MET 45:
HB2/HB3 MET 45: 35/0 conformations, assigned as input.
HG2/HG3 MET 45: 14/21 conformations, unassigned.
Delete or comment out those lines in stereofound.cya, which you think correspond to incorrectly derived stereospecific assignments.
In subsequent steps it is recommended to swap the labels in the original source containing chemical shift assignments: the atom list if using XEASY, or CARA repository if using CARA.
- GridSearch.cya: CYANA script to run FOUND module
- CALC.cya: CYANA script to run manual calculation with local constraints