Mass Spectrometry: Difference between revisions
No edit summary |
|||
| (16 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
== '''Compare Mass Spectrometry m/z with Calculated Protein Mass''' == | == '''Compare Mass Spectrometry m/z with Calculated Protein Mass''' == | ||
=== Mass Spectrometry Results from | === Mass Spectrometry Results from Rutger's University<br> === | ||
<br> | <br> | ||
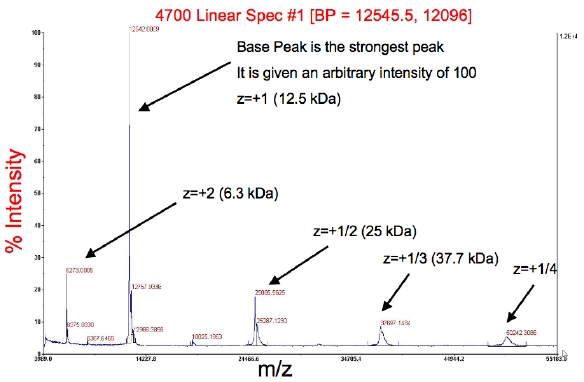
At Rutgers University, spectra are collected on a MALDI-TOF/TOF ([http://cabm-ms.cabm.rutgers.edu/instruments.html ABI-MDS SCIEX 4800]) in single TOF mode. Samples are prepared by mixing 1 ul of the final protein NMR sample with 10 ul of sinapinic acid (SA) matrix solution<!--StartFragment--> <font face="Verdana, Helvetica, Arial"><span style="font-size: 12px;">(10 mg/ml SA in 50% ACN/50% 0.1% TFA).</span></font> | |||
<font face="Verdana, Helvetica, Arial"><br></font> | |||
Results: | |||
*The peaks should correspond to your full-length protein mass. | *The peaks should correspond to your full-length protein mass. | ||
*The red information across the top is the base peak, BP (the peak with the greatest intensity), followed by the intensity | *The red information across the top is the base peak, BP (the peak with the greatest intensity), followed by the intensity | ||
*The other peaks are typically m/z (mass-to-charge ratio) with z = 2+, 1/2+, 1/3+, etc. | *The other peaks are typically m/z (mass-to-charge ratio) with z = 2+, 1/2+, 1/3+, etc. <!--EndFragment--> | ||
<br> | <br>At Rutger's mass spectrometry data is collected for each NC5 sample, NC sample,and unlabelled sample. Error bars for this measurement are typically < 25 Da, but can be more. | ||
<br>Some reasons that the MS mass may not match your calculated mass are: | <br>Some reasons that the MS mass may not match your calculated mass are: | ||
| Line 31: | Line 34: | ||
=== N-terminal Methionine Cleavage === | === N-terminal Methionine Cleavage === | ||
<br>The propensity for cleavage has been analyzed in a | <br>The propensity for cleavage has been analyzed in a 2006 bioinformatics paper by Frottin ''et al.'':<ref>[http://www.ncbi.nlm.nih.gov/pubmed/16963780?dopt=Abstract F. Frottin, A. Martinez, P. Peynott, M. Sanghamitra, R. C. Holz, C. Giglione, T. Meinnel, Mol Cell Proteomics, 2006, 5, 2336-49]</ref> | ||
<br>In summary, N-Met cleavage is: | |||
*Very likely if the second residue is: A, C, G, or | *Very likely if the second residue is: A, C, G, P or S | ||
*Not likely if the second residue is V or T. | *Not likely if the second residue is V or T. | ||
*There are other more complicated influences from the 3rd and 4th residue. | *There are other more complicated influences from the 3rd and 4th residue. | ||
| Line 49: | Line 52: | ||
=== Calculate the expected molecular weight for the NC5 and NC sample === | === Calculate the expected molecular weight for the NC5 and NC sample === | ||
<br> | |||
[http://www.scripps.edu/~cdputnam/protcalc.html Protein Calculator] is a nice tool that allows to calculate molecular mass of isotopically labeled proteins | [http://www.scripps.edu/~cdputnam/protcalc.html Protein Calculator] is a nice tool that allows to calculate molecular mass of isotopically labeled proteins | ||
<br>Another way to calculate the molecular weight for a protein is using the [http://ca.expasy.org/tools/protparam.html ExPASY ProtParam] tool. Just paste your sequence into the web form. The atomic composition at the bottom of the results can be used to correct for the isotopic enrichment as in the examples below. | <br>Another way to calculate the molecular weight for a protein is using the [http://ca.expasy.org/tools/protparam.html ExPASY ProtParam] tool. Just paste your sequence into the web form. The atomic composition at the bottom of the results can be used to correct for the isotopic enrichment as in the examples below. | ||
=== Examples === | === Examples === | ||
| Line 74: | Line 79: | ||
So, the calculated MWs are:<br> Unlabeled: 11726 Da<br> NC5: 11726 + (165 Da for 15N) + (0.05 x 505 Da for 13C) = 11916 Da <br> NC: 11726 + (165 Da for 15N) + (505 Da for 13C) = 12396 Da<br><br>From Spine, the MS results are:<br> LBa: 11592 Da (-134 Da from expected)<br> NC5a: 11779 Da (-137 Da from expected)<br> NC5c: 11775 Da (-141 Da from expected)<br> NCa: 12247 Da (-149 Da from expected)<br><br>Since unlabelled Met is 131 Da (and for NC5 is 134, and NC is 137 Da), it seems likely that the N-terminal Met is cleaved.<br><br>Note. Terminator3 does not predict N-terminal Met cleavage, even though the second residue, Ala, makes it possible. | So, the calculated MWs are:<br> Unlabeled: 11726 Da<br> NC5: 11726 + (165 Da for 15N) + (0.05 x 505 Da for 13C) = 11916 Da <br> NC: 11726 + (165 Da for 15N) + (505 Da for 13C) = 12396 Da<br><br>From Spine, the MS results are:<br> LBa: 11592 Da (-134 Da from expected)<br> NC5a: 11779 Da (-137 Da from expected)<br> NC5c: 11775 Da (-141 Da from expected)<br> NCa: 12247 Da (-149 Da from expected)<br><br>Since unlabelled Met is 131 Da (and for NC5 is 134, and NC is 137 Da), it seems likely that the N-terminal Met is cleaved.<br><br>Note. Terminator3 does not predict N-terminal Met cleavage, even though the second residue, Ala, makes it possible. | ||
=== References === | |||
<references /> | |||
Latest revision as of 18:29, 1 October 2015
Compare Mass Spectrometry m/z with Calculated Protein Mass
Mass Spectrometry Results from Rutger's University
At Rutgers University, spectra are collected on a MALDI-TOF/TOF (ABI-MDS SCIEX 4800) in single TOF mode. Samples are prepared by mixing 1 ul of the final protein NMR sample with 10 ul of sinapinic acid (SA) matrix solution (10 mg/ml SA in 50% ACN/50% 0.1% TFA).
Results:
- The peaks should correspond to your full-length protein mass.
- The red information across the top is the base peak, BP (the peak with the greatest intensity), followed by the intensity
- The other peaks are typically m/z (mass-to-charge ratio) with z = 2+, 1/2+, 1/3+, etc.
At Rutger's mass spectrometry data is collected for each NC5 sample, NC sample,and unlabelled sample. Error bars for this measurement are typically < 25 Da, but can be more.
Some reasons that the MS mass may not match your calculated mass are:
- The MS results might have large error because of instrument or calibration problems (unlikely).
- The sequence in the database is incorrect.
- There are mutations in your protein (very common).
- The protein has covalent modifications.
- Protein degradation occurs.
- The N-terminal methionine is cleaved in the E.coli due to N-terminal methione excision (NME) by methione aminopeptidases (MAPs).
Example Mass Spectrum of TaR80B
http://spine.nesg.org/batch.cgi?id=TaR80B-21.2-NCa-GF (password protected)
N-terminal Methionine Cleavage
The propensity for cleavage has been analyzed in a 2006 bioinformatics paper by Frottin et al.:[1]
In summary, N-Met cleavage is:
- Very likely if the second residue is: A, C, G, P or S
- Not likely if the second residue is V or T.
- There are other more complicated influences from the 3rd and 4th residue.
- Overexpressed proteins are less likely to be cleaved than less highly expressed proteins.
They developed a bioinformatics tool for prediction called TermiNator (also available through ExPASY):
Use these Terminator options for proteins overexpressed in E.coli:
- expressed in: Prokaryote (Eubacteria or Archea)
- Eubacteria
- Recombinant, plasmid-encoded, overexpressed gene
- LPR cleavage (leader peptide cleavage) – No
- Paste your sequence
Calculate the expected molecular weight for the NC5 and NC sample
Protein Calculator is a nice tool that allows to calculate molecular mass of isotopically labeled proteins
Another way to calculate the molecular weight for a protein is using the ExPASY ProtParam tool. Just paste your sequence into the web form. The atomic composition at the bottom of the results can be used to correct for the isotopic enrichment as in the examples below.
Examples
FR629A example (no N-met cleavage)
MGHHHHHHSHGKSDFIKVNVSNSHNDAVAFEVKLAKDLTVAQLKTKLEILTGGCAGTMKVQVFKGDTCVSTMDNNDAQLGYYANSDGLRLHVVDS
FR629A has the following ProtParam results:
Molecular weight: 10343
Atomic composition:
Carbon C 447
Nitrogen N 133
So, the calculated MWs are:
Unlabeled: 10343 Da
NC5: 10343 + (133 Da for 15N) + (0.05 x 447 Da for 13C) = 10500 Da
NC: 10343 + (133 Da for 15N) + (447 Da for 13C) = 10923 Da
From Spine, the MS results are:
NC5a: 10488 Da (-11 Da from expected)
NC5c: 10497 Da (-3 Da from expected)
NCa: 10915 Da (+ 8 Da from expected)
This is a good match. Terminator3 does not predict N-terminal Met cleavage, even though the second residue, Gly, makes it possible.
MbR242E example (with N-Met cleavage)
MAGQSDRKAALLDQVARVGKALANGRRLQILDLLAQGERAVEAIATATGMNLTTASANLQALKSGGLVEARREGTRQYYRIAGEDVARLFALVQVVADEHLEHHHHHH
MbR242E has the following ProtParam results:
Molecular weight: 11726
Atomic composition:
Carbon C 505
Nitrogen N 165
So, the calculated MWs are:
Unlabeled: 11726 Da
NC5: 11726 + (165 Da for 15N) + (0.05 x 505 Da for 13C) = 11916 Da
NC: 11726 + (165 Da for 15N) + (505 Da for 13C) = 12396 Da
From Spine, the MS results are:
LBa: 11592 Da (-134 Da from expected)
NC5a: 11779 Da (-137 Da from expected)
NC5c: 11775 Da (-141 Da from expected)
NCa: 12247 Da (-149 Da from expected)
Since unlabelled Met is 131 Da (and for NC5 is 134, and NC is 137 Da), it seems likely that the N-terminal Met is cleaved.
Note. Terminator3 does not predict N-terminal Met cleavage, even though the second residue, Ala, makes it possible.