Resonance Assignment/Abacus/FMCGUI commands: Difference between revisions
No edit summary |
|||
| (172 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
<div> </div><div> </div> | <div> </div><div> </div> | ||
| | ||
== | == Project Menu == | ||
| | ||
=== <span>'''Project>New'''</span> === | === <span>'''Project>New'''</span> === | ||
<div><span>:Start a new project | <div>'''<span>::</span>'''<span> Start a new project </span></div><div></div><div> User have to provide a name of the project PROJECTNAME,and to select a directory that will host the project. The project root directory with the same name PROJECTNAME is created. </div><div><span> </span></div> | ||
=== '''<span>Project>Load</span>''' === | === '''<span>Project>Load</span>''' === | ||
<div>: Continue to work | <div>:: Continue to work with previously saved project.</div><div> </div><div>User have to select file PROJECTNAME.prj in the directory PROJECTNAME, where PROJECTNAME is the name of the root directory of the project.</div><div> </div> | ||
=== '''<span>Project>Save</span>''' === | === '''<span>Project>Save</span>''' === | ||
<div></div><div> : | <div></div><div> :: Save the current state of the project</div><div> </div><div>What is currently in the computer memory is saved in the file PROJECTNAME.prj located in the root directory of the project. </div><div> </div> | ||
=== '''<span>Poject>Quit</span>''' | === '''<span>Poject>Quit</span>''' === | ||
<div> </div><div>:: Save the current state of the project and quit.</div><div> </div> | |||
== Data Menu<br> == | |||
<div>The DATA section serves to load&save the data (such as protein sequence and peak lists). Since there are different formats of data-files that could be loaded in memory or saved on disk, one can use this section as format converter as well.</div><div> </div> | |||
=== '''<span>Data>Protein Sequence>Load</span>''' === | |||
<div></div><div>:: Load [[FMCGUI objects#Protein_sequence|protein sequence]] into memory.</div><div> </div><div>''The input formats'': </div><div><span> </span> <span> 1-letter code ([[Protein Sequence format|fasta format]]); </span></div><div><span> 3-letter code ([[Protein Sequence format|standard format]]).</span></div><div> </div><div>User have to select the file with sequence and to specify the first residue ID in case it is not specified in the input file. If there is His-tag in the sequence file, it is recommended to set the first residue ID to a negative number such that the first residue of a protein has ID of 1. </div><div> </div> | |||
| === '''<span>Data>Protein Sequence>Save as</span>''' === | ||
<div></div><div>:: Save protein sequence in file on disk.</div><div> </div><div>''The output formats'':</div><div><span> 1-letter code ([[Protein Sequence format|fasta format]]);</span></div><div><span> 3-letter code ([[Protein Sequence format|standard format]], for cyana)</span></div><div><span> 3-letter code (for AutoStructure);</span></div><div><span> 3-letter code (for RCI);</span></div><div> <br></div> | |||
=== '''<span>Data>N15 NOESY></span>''' === | === '''<span>Data>N15 NOESY></span>''' === | ||
<div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. <div>Referencing is not required for NOESY peak list </div><div></div><div></div><div>''Input and output formats'': </div><div><span> [[Peak Lists format|Sparky]]</span></div><div><span> [[Peak Lists format|Xeasy]]</span></div><div><span> [[Peak Lists format|Standard]]</span></div><div><span> </span></div></div> | |||
| === '''<span>Data>C13 NOESY></span>''' === | ||
<div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. <div>Referencing is not required for NOESY peak list </div><div>''Input and output formats'': </div><div><span> [[Peak Lists format|Sparky]]</span></div><div><span> [[Peak Lists format|Xeasy]]</span></div><div><span> [[Peak Lists format|Standard]]</span></div> | |||
<span> </span> | |||
<br></div> | |||
=== '''<span>Data>Arom NOESY></span>''' === | |||
<div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. | |||
<div>Referencing is not required for NOESY peak list </div><div>''Input and output formats'': </div><div><span> [[Peak Lists format|Sparky]]</span></div><div><span> [[Peak Lists format|Xeasy]]</span></div><div><span> [[Peak Lists format|Standard]]</span></div> | |||
<span> | |||
</span> | |||
=== '''<span>Data>N15 HSQC></span>''' === | === '''<span>Data>N15 HSQC></span>''' === | ||
<div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. <div></div><div>This peak list should be referenced.</div><div> </div><div>''Input and output formats'': </div><div><span> [[Peak Lists format|Sparky]]</span></div> | |||
<span> | |||
</span> | |||
| === '''<span>Data>C13 HSQC></span>''' === | ||
<div></div> <div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. <div><br></div> <div>This peak list should be referenced.<br></div> <div> </div> <div>''Input and output formats'': </div> <div><span> [[Peak Lists format|Sparky]]</span></div></div> | |||
=== '''<span>Data>HNCA></span>''' === | === '''<span>Data>HNCA></span>''' === | ||
<div></div> | <div></div> <div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. <div><br></div> <div>It is recommended this peak list to be referenced.<br></div> <div> </div> <div>''Input and output formats'': </div> <div><span> [[Peak Lists format|Sparky]]</span></div> <div><span> [[Peak Lists format|Xeasy]]</span></div> <div><span> [[Peak Lists format|Standard]]</span></div> | ||
=== '''<span>Data>HNCO></span>''' | <span> | ||
</span> | |||
=== '''<span>Data>HNCO></span>''' === | |||
<div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. | |||
<div><br></div> <div>It is not required this peak list to be referenced.<br></div> <div> </div> <div>''Input and output formats'': </div> <div><span> [[Peak Lists format|Sparky]]</span></div> <div><span> [[Peak Lists format|Xeasy]]</span></div> <div><span> [[Peak Lists format|Standard]]</span></div> | |||
=== '''<span>Data>CBCACONHN></span>''' === | === '''<span>Data>CBCACONHN></span>''' === | ||
<div></div> | <div></div> | ||
<div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. | |||
<div><br></div> <div>It is not reauired this peak list to be referenced.<br></div> <div> </div> <div>''Input and output formats'': </div> <div><span> [[Peak Lists format|Sparky]]</span></div> <div><span> [[Peak Lists format|Xeasy]]</span></div> <div><span> [[Peak Lists format|Standard]]</span></div> | |||
=== '''<span>Data>HBHACONH></span>''' === | === '''<span>Data>HBHACONH></span>''' === | ||
<div> | <div>:: Load or save [[FMCGUI objects#Peak_list|peak list]]. <div><br></div> <div>This peak list should be referenced.<br></div> <div> </div> <div>''Input and output formats'': </div> <div><span> [[Peak Lists format|Sparky]]</span></div> <div><span> [[Peak Lists format|Standard]]</span></div> <span> | ||
</span></div> | |||
=== '''<span>Data>Tolerances</span>''' === | === '''<span>Data>Tolerances</span>''' === | ||
<div></div><div>: | <div></div> <div>:: Set tolerances for chemical shift matching in different spectral dimensions.</div><div> </div> | ||
There are six tolerances to set up:<br> | |||
NX - tolerance for matching resonances in N15 dimension | |||
CX - tolerance for matching resonances in C13 dimension<br> | |||
HN - tolerance for matching resonances in HN direct dimension<br> | |||
HC - tolerance for matching resonances in HC direct dimension<br> | |||
Hi - tolerance for matching resonances in H indirect dimension<br> | |||
Ci - tolerance for matching resonances in C13 indirect dimension<br> | |||
<br> | <br> | ||
<br> | <br> | ||
== | == Fragment menu == | ||
=== '''<span>Fragment>Load>assigned</span>''' === | === '''<span>Fragment>Load>assigned</span>''' === | ||
<div></div><div><span>: | <div></div><div><span>:: Load assigned chemical shifts (spin-systems) in memory. </span></div><div> </div><div>''Prerequisites:''</div><div>''<span> </span>''♦ ''<span> </span>''Loaded sequence</div><div> </div><div>''Input formats'': </div><div><span> </span>♦ <span> [[Spin systems format|assigned AA-fragments in standard format]]</span></div><div><span> </span>♦ <span> [[Spin systems format|CYANA chemical shift file (prot-file)]]</span></div><div> </div> | ||
=== '''<span>Fragment>Load>PB fragments</span>''' === | === '''<span>Fragment>Load>PB fragments</span>''' === | ||
<div></div><div><span>: | <div></div><div><span>:: Load unassigned spin-systems ([[Introduction to ABACUS#PB_fragment|PB fragments]]) in memory.</span></div><div> </div><div>''Input format'' :</div><div><span> </span>♦<span> [[Spin systems format|PB fragments in standard format]].</span></div><div> </div><div> </div> | ||
=== '''<span>Fragment>Save>PB fragments</span>''' === | === '''<span>Fragment>Save>PB fragments</span>''' === | ||
<div><span>:: Save PB-fragments in a file on disk.</span></div><div> </div><div>All PB fragments are saved <span> [[Spin systems format|in standard format]]. </span></div><div> </div><div>The name of the saved file and it’s location are specified by user. </div><div>There are 3 options to save PB-fragments in the file:<span> </span></div> | |||
#<span> In the order of fragments index, that is in the order by which fragments are stored in memory;</span> | |||
#<span> In the order of fragments [[FMCGUI objects#User_ID|user ID]]</span> | |||
#<span> In the order of fragments [[FMCGUI objects#Assignment_ID|assignment ID]], ''A_id''. In this case two files are saved. One file, with user specified name 'user_name', contains only fragments assigned to protein sequence positions, that is to positions with residue ID of >= 1. The second file, with the name 'user_name_na', contains all not assigned fragments (that is fragments with ''A_id'' = -99).</span> | |||
<br> | |||
=== '''<span>Fragment>Save>cyana</span>''' === | === '''<span>Fragment>Save>cyana</span>''' === | ||
<div></div><div><span>: | <div></div><div><span>:: Save assigned chemical shifts (that is fragments with [[FMCGUI_objects#Assignment_ID|assignment ID]] > 0 ) in [[Spin_systems_format|CYANA format]].</span></div><div> </div> | ||
=== '''<span>Fragment>Save>bmrb</span>''' === | === '''<span>Fragment>Save>bmrb</span>''' === | ||
<div></div><div><span>: | <div></div><div><span>:: </span><span>Save assigned chemical shifts (that is fragments with [[FMCGUI objects#Assignment_ID|assignment ID]] > 0 )</span><span> in the format suitable for BMRB deposition (star2.1)</span></div><div> </div> | ||
=== '''<span>Fragment>Save>talos</span>''' === | === '''<span>Fragment>Save>talos</span>''' === | ||
<div></div><div><span> : | <div></div><div><span> :: </span><span>Save assigned chemical shifts (that is fragments with [[FMCGUI objects#Assignment_ID|assignment ID]] > 0 )</span><span> in the format suitable for TALOS/CS-Rosetta;</span></div><div> </div><div></div> | ||
=== '''<span><span | |||
<div></div><div><span>: | === '''<span><span style="display: none;" id="1259364396632S"> </span><span style="display: none;" id="1259364443776S"> </span>Fragment>Save>abacus</span>''' === | ||
<div></div><div><span>:: </span><span>Save assigned chemical shifts (that is fragments with [[FMCGUI objects#Assignment_ID|assignment ID]] > 0 )</span><span> in the format suitable for BACUS;</span></div><div> </div><div> </div> | |||
=== '''Fragment>Create>fawn''' === | === '''Fragment>Create>fawn''' === | ||
<div></div><div>: | <div></div><div>:: Create and evaluate ''b''PB fragments</div><div> </div><div>''Prerequisites'': </div> | ||
♦ loaded in memory <u>referenced</u> peak lists of CBCA(CO)HN, HBHA(CO)HN, N15HSQC, and HNCA spectra; | |||
<div> | <div> ♦ Specified tolerances.</div><div><span> </span></div><div>There are two steps in executing this command.</div><div></div> | ||
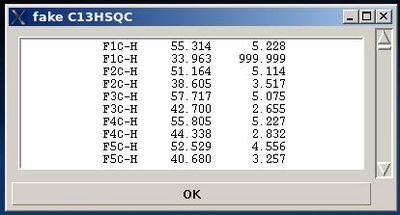
#On the first step, a fake C13HSQC peak list is created and shown in the popped up window “fake C13HSQC” .<br> | |||
<div>[[Image:Fmcgui fig2 2.jpg|thumb|right|400px]] </div><div></div><div></div><div>User can use the information shown in the main FMCGUI window and check /edit the list in the entry section of “fake C13HSQC” window accordingly. </div><div>Pressing OK will result in loading the peak list from the entry window into memory as C13HSQC peak list.</div><div> </div> | |||
<br> | |||
<br> | |||
<br> | |||
<br> | |||
<br> | |||
<br> 2. On the second step, a number of ''b''PB fragments corresponding to 20 different amino acid types are generated from user-identified spin-systems. | |||
<br> | |||
Each generated ''b''PB fragment is evaluated by a score S(T) that measure how good the spin-system chemical shifts match corresponding statistical chemical shifts derived from BMRB database (see Figure 1.2). The ''b''PB fragment with highest score is selected to form a list of bPB-fragments. | |||
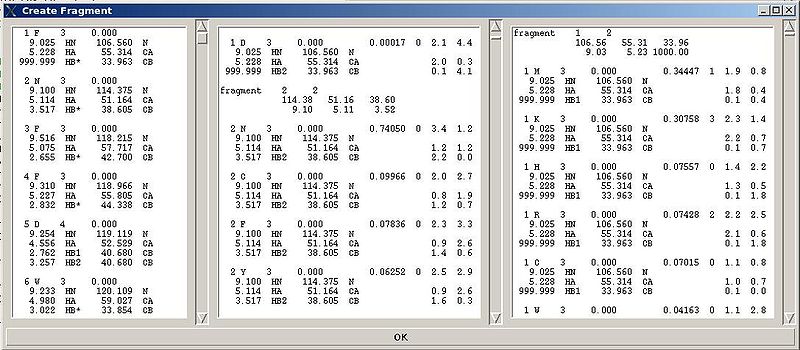
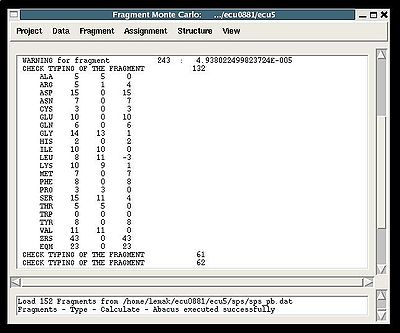
<div>The warning messages of the command are shown in the main FMCGUI window (see for example, Figure 2.4).</div><div></div><div></div><div></div><div></div><div></div><div></div><div></div><div></div><div></div><div></div><div></div><div></div><div>In addition to that, a new window ‘Create Fragment’ pops up:<br> </div><div>.</div><div></div> <div></div><div></div><div></div><div>[[Image:Fmcgui fig2.3.jpg|thumb|center|800px]]</div><div>"Create Fragment" window consists of three sections. The left sections contains suggested bPB fragments, while the other sections contains two reports of fragments scoring with both C and H resonances and with only C resonances, respectively. Following the warning messages shown in the main FMCGUI window (see Figure 2.4), user can accept/modify generated ''b''PB fragments. Alternatively, when ‘poor’ ''b''PB fragments are present, user can go back to spectra, fix the pick lists accordingly, and repeat the fragment generation again.</div><div>User-approved ''b''PB fragments will be loaded in the memory by pressing OK button.</div><div> </div><div> <br></div><div>''Results:''</div><div><span><span> ♦ </span></span>C13HSQC peak list loaded in memory</div><div><span><span> ♦ </span></span>bPB fragments are loaded in memory</div><div> </div> | |||
=== Fragment>Create>abacus === | |||
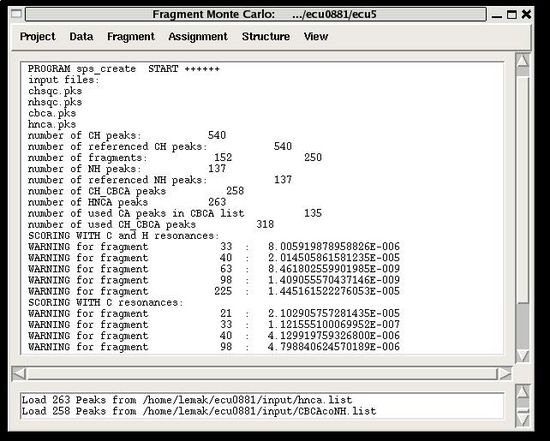
<div>:: Create and avaluate PB fragments.</div><div> </div><div></div><div></div><div>''Prerequisites'': </div><div> ♦ Loaded in memory <u>referenced</u> C13HSQC, N15HSQC, and HNCA peak lists and <u>not referenced</u> CBCA(CO)HN peak list.</div><div> ( As an option, HNCA peak list could be not referenced as well)</div><div> ♦ Specified tolerances.</div><div> <br></div><div><div> </div><div>''Results:''</div><div><span> <span> </span></span>♦ PB fragments are loaded in memory</div><div> <br></div></div><div>A number of PB fragments corresponding to 20 different amino acid types are generated from user-identified spin-systems. Each generated PB fragment is evaluated by a score ''S'' (see Figure 1.2) that measure how good the spin-system chemical shifts match corresponding statistical chemical shifts derived from BMRB database. The PB fragment with highest score is selected to form a list of PB fragments.</div><div>Spin-system which have S<sub>max</sub> less than 10<sup>-4</sup> are reported in the warning lines shown in the main FMCGUI window: </div> | |||
==== '''Figure 2.4''' ==== | |||
<div> | |||
[[Image:Fmcgui fig2.4.jpg|thumb|center|550px]] | |||
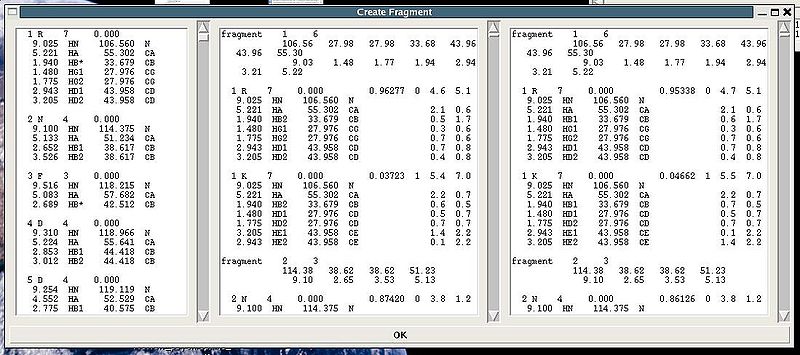
</div><div><br>Following these warnings user can check and modify generated PB fragments in the left section of “Create Fragment’ window. </div><div>.</div><div></div><div>[[Image:Fmcgui fig2.5.jpg|thumb|center|800px]]</div><div></div><div><div></div><div>"Create Fragment" window consists of three sections. The left sections contains suggested PB fragments, while the other two sections contain report of fragment's scoring with both C and H atom resonances and with only C atom resonances, respectively. Following the warning messages shown in the main FMCGUI window (see Figure 2.4), user can accept/modify generated ''b''PB fragments. </div><div>Pressing OK button, user-approved PB fragments shown in the left section of "Create Fragment" window will be loaded in the memory.</div><div>Alternatively, when ‘poor’ PB fragments are present, user can go back to spectra, fix the pick lists accordingly, and repeat the fragment generation again.</div><br></div> | |||
=== '''Fragment>Type>Calculate>''' === | === '''Fragment>Type>Calculate>''' === | ||
<div>:: Probabilistic typing of bPB (fawn) or PB (abacus) fragments <div> <br></div><div>''Prerequisites'': </div><div> ♦ loaded in memory protein sequence</div><div> ♦ loaded in memory PB-fragments</div><div> ♦ specified tolerances.</div><div> </div><div>''Results:''</div><div> ♦ Fragment typing probabilities <span>are calculated and loaded in memory.</span></div><div> <br></div><div>[[Image:Fmcgui fig2.6.jpg|thumb|left|400px]] </div><div> </div><div></div><div></div><div>The main FMCGUI window displays (see Figure ):</div> | |||
*<span><span> </span></span>the summary table that shows how many fragments of each AA-residue type are expected and how many fragments were actually recognized by the typing script | |||
*<span><span> </span></span>warning messages that suggest user to check and possibly modify typing manually of some fragments manually (see command {Fragment>Type>fix}) | |||
<div> </div><div> </div><div> </div><div> </div><div> <br> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div><br> </div></div> | |||
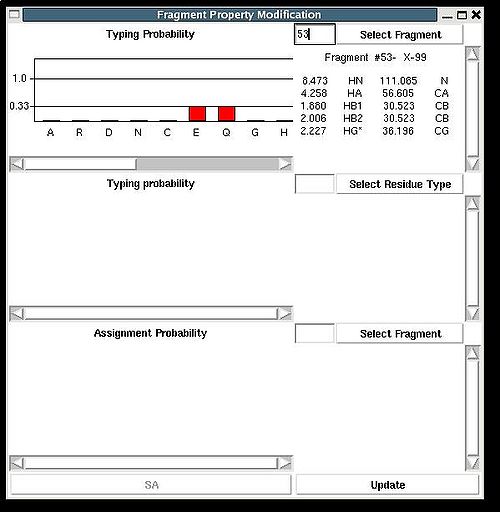
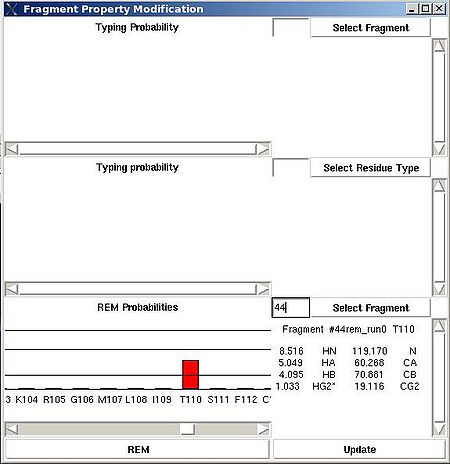
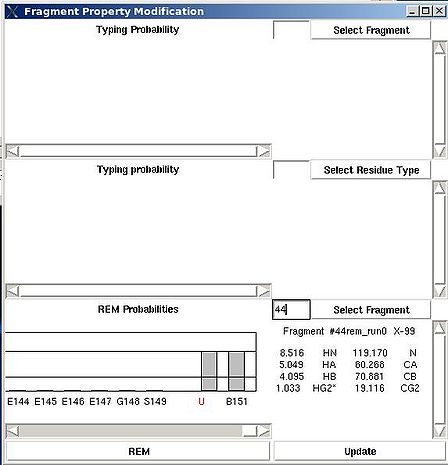
=== <span>Fragment>Type>fix</span> === | === <span>Fragment>Type>fix</span> === | ||
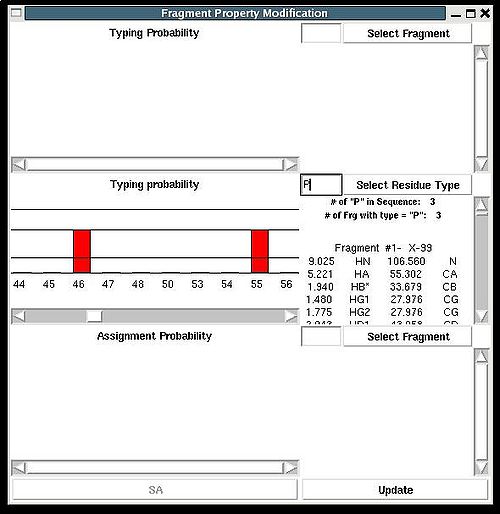
<div>''':''' | <div>''':''': Modify typing probabilities manually.<br></div><div> <br></div><div>''Prerequisites'': </div><div> ♦ loaded in memory protein sequence</div><div> ♦ loaded in memory PB fragments</div><div> </div><div>The command opens "Fragment Property Modification" (FPM) window (see Figure). </div><div> </div><div> [[Image:Fmcgui fig2.7b.jpg|thumb|center|500px]] </div><div> </div><div>This window has 3 sections. </div><div>In the top section of FPM window user can select fragment user ID, ''U_id''. </div><div> </div><div> [[Image:Fmcgui fig2.7a.jpg|thumb|center|500px]] </div><div> </div><div>Then typing probabilities <span>T<sup>t</sup>(''U_id'') for all 20 amino acid types ''t'' will be shown on the graph. The chemical shifts of the fragments and its assignment status (''A_id'') are shown as well. User can modify typing probabilities of the fragment </span><span>''U_id .''</span><span> Selecting amino acid types on the graph using right mouse button and pressing ‘Update’ button on the bottom of FPM window will set probabilities </span><span>T<sup>t</sup>(''U_id'') </span><span>corresponding to the selected amino acid types to the same non-zero value (such that the normalization condition is satisfied).</span></div><div> <br></div><div>In the middle section of FPM window user can select amino acid residue type ''t1 ''( for example, on the following Figure, ''t1 = ''P ). </div><div> </div><div> [[Image:Fmcgui fig2.8.jpg|thumb|center|500px]] </div><div> <br>Then the graph will show typing probabilities T<sup>t1</sup>(f) <span />that correspond to the selected residue type ''t1'' for all available fragments IDs ''f'' . </div><div>Selecting a particular fragment ''U_id'' on the graph using right mouse button (the color of ''U_id'' will be changed to red) and then pressing “Update” button will set the probability T<sup>t1</sup>(''U_id'' )<span> to 1.</span></div><div><span> </span>Selecting a particular fragment ''U_id'' on the graph using left mouse button (the color of ''U_id ''wil be changed to blue) and then pressing “Update” button will set the probability T<sup>t1</sup>(''U_id'' )<span> </span><span>to 0.</span></div><div> </div> | ||
=== '''Fragment>Expected Peaks>''' === | === '''Fragment>Expected Peaks>''' === | ||
<div>''':''' | <div>'''::''' Generate lists of peaks expected from covalent structure of PB fragments.</div><div> </div><div> | ||
''Prerequisites'': | |||
♦ protein sequence loaded in memory | |||
♦ PB-fragments loaded in memory | |||
| |||
Expected peak lists of the following spectra could be generated: N15-NOESY, C13-NOESY, H(C)CH-TOCSY, (H)CCH-TOCSY, | |||
N15-HSQC, and C13-HSQC. Generated peak list is saved to the file on disk in SPARKY format. <br> | |||
</div> | |||
=== '''Fragment>Modify assigned''' === | === '''Fragment>Modify assigned''' === | ||
<div>''': ''' | <div>'''::''' Correct assigned fragments.</div><div> </div><div>''Prerequisites'': </div> | ||
== | ♦ loaded in memory protein sequence | ||
♦ loaded in memory PB-fragments | |||
♦ loaded HNCO, CBCACONH, and HNCA peak lists. | |||
♦ specified tolerances. | |||
<div> <br></div><div> ''Resuls:''</div><div> ♦ CO chemical shifts are assigned </div><div> ♦ chemical shift names are corrected for assigned PB-fragments (the fragments with A_id > -99 )</div><div> <br></div> | |||
== Assignment menu == | |||
<div> | <div> | ||
=== '''Assignment>Contacts>HNCA''' === | === '''Assignment>Contacts>HNCA''' === | ||
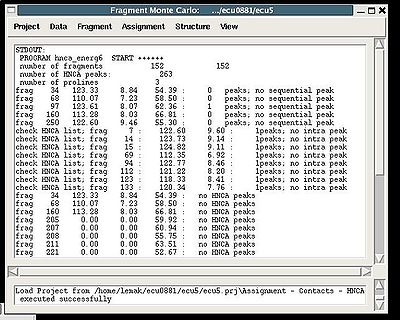
''':''' | ''':''': Calculate <span> fragments contact map C<sub>HNCA</sub></span> <br> | ||
< | <div> <br></div><div>''Prerequisites'': </div><div> ♦ PB-fragments are loaded in memory</div><div> ♦ typing probabilities <span>are calculated </span></div><div> ♦ HNCA peak list (recommended to be referenced) is loaded in memory</div><div> ♦ tolerances are specified</div><div> </div><div>''Results:''</div><div> ♦ Contact map C<sub>HNCA</sub> <span>is calculated and loaded in memory</span></div><div> <br> </div><div>During the execution of this command the loaded HNCA peak list is validated. The validation results are shown in the project main window.</div><div> </div><div>[[Image:Fmcgui fig2.9b.jpg|thumb|left|400px]] </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> </div><div> <br></div><div></div><div>User have to check suspicious peaks going back to the spectra and correct the input peak list if it is needed. </div><div> <br></div><div> </div><div> </div> | ||
=== '''Assignment>Contacts>NOE>fawn''' === | === '''Assignment>Contacts>NOE>fawn''' === | ||
<div>''':''' | <div>''':: ''' Calculate <span> fragment contact map C<sub>NOE_F</sub>.</span></div><div> </div><div>''Prerequisites'': </div><div> ♦ PB-fragments are loaded in memory;</div><div> ♦ typing probabilities <span>are calculated; </span></div><div> ♦ N15-NOESY peak list is loaded in memory.</div><div> ♦ tolerances are specified.</div><div> <br>''Results:''<br> </div><div> ♦ contact map C<sub>NOE_F</sub> <span>is calculated using N15-NOESY peak list and loaded in memory</span></div><div> </div><div> </div> | ||
=== '''Assignment>Contacts>NOE>abacus''' === | === '''Assignment>Contacts>NOE>abacus''' === | ||
<div>''':''' | <div>''':''': Calculate both C<sub>NOE_B</sub><span> and C<sub>NOE_F</sub> contact maps.</span></div><div> </div><div>''Prerequisites'': </div><div> ♦ PB-fragments are loaded in memory</div><div> ♦ typing probabilities <span>are calculated </span></div><div> ♦ N15-NOESY and C13-NOESY peak lists are loaded in memory</div><div> ♦ tolerances are specified</div><div> <br></div><div>''Results:''</div><div> ♦ C<sub>NOE_F</sub> contact map <span>is calculated using only N15_NOESY peak list </span></div><div><span> </span>♦ C<sub>NOE_B</sub><span> contact map is calculated using both N15_NOESY and C13_NOESY peak lists that are interpreted by BACUS procedure</span></div><div><span> </span>♦<span> Both calculated maps are loaded in memory</span></div><div><span> </span></div><div> </div><div> </div><div> </div><div> </div> | ||
=== '''Assignment>Calculate Probabilities>SA''' === | === '''Assignment>Calculate Probabilities>SA''' === | ||
<div>'''::''' Calculate <span>assignment probabilities </span></div><div> </div><div>''Prerequisites'': </div><div> & | |||
Latest revision as of 23:33, 5 January 2010
Project Menu
Project>New
Project>Load
Project>Save
Poject>Quit
Data Menu
Data>Protein Sequence>Load
Data>Protein Sequence>Save as
Data>N15 NOESY>
Data>C13 NOESY>
Data>Arom NOESY>
Data>N15 HSQC>
Data>C13 HSQC>
Data>HNCA>
Data>HNCO>
Data>CBCACONHN>
Data>HBHACONH>
Data>Tolerances
There are six tolerances to set up:
NX - tolerance for matching resonances in N15 dimension
CX - tolerance for matching resonances in C13 dimension
HN - tolerance for matching resonances in HN direct dimension
HC - tolerance for matching resonances in HC direct dimension
Hi - tolerance for matching resonances in H indirect dimension
Ci - tolerance for matching resonances in C13 indirect dimension
Fragment>Load>assigned
Fragment>Load>PB fragments
Fragment>Save>PB fragments
- In the order of fragments index, that is in the order by which fragments are stored in memory;
- In the order of fragments user ID
- In the order of fragments assignment ID, A_id. In this case two files are saved. One file, with user specified name 'user_name', contains only fragments assigned to protein sequence positions, that is to positions with residue ID of >= 1. The second file, with the name 'user_name_na', contains all not assigned fragments (that is fragments with A_id = -99).
Fragment>Save>cyana
Fragment>Save>bmrb
Fragment>Save>talos
Fragment>Save>abacus
Fragment>Create>fawn
♦ loaded in memory referenced peak lists of CBCA(CO)HN, HBHA(CO)HN, N15HSQC, and HNCA spectra;
- On the first step, a fake C13HSQC peak list is created and shown in the popped up window “fake C13HSQC” .
2. On the second step, a number of bPB fragments corresponding to 20 different amino acid types are generated from user-identified spin-systems.
Each generated bPB fragment is evaluated by a score S(T) that measure how good the spin-system chemical shifts match corresponding statistical chemical shifts derived from BMRB database (see Figure 1.2). The bPB fragment with highest score is selected to form a list of bPB-fragments.
Fragment>Create>abacus
Figure 2.4
Following these warnings user can check and modify generated PB fragments in the left section of “Create Fragment’ window.
Fragment>Type>Calculate>
- the summary table that shows how many fragments of each AA-residue type are expected and how many fragments were actually recognized by the typing script
- warning messages that suggest user to check and possibly modify typing manually of some fragments manually (see command {Fragment>Type>fix})
Fragment>Type>fix
Then the graph will show typing probabilities Tt1(f) <span />that correspond to the selected residue type t1 for all available fragments IDs f .
Fragment>Expected Peaks>
Prerequisites:
♦ protein sequence loaded in memory
♦ PB-fragments loaded in memory
Expected peak lists of the following spectra could be generated: N15-NOESY, C13-NOESY, H(C)CH-TOCSY, (H)CCH-TOCSY,
N15-HSQC, and C13-HSQC. Generated peak list is saved to the file on disk in SPARKY format.
Fragment>Modify assigned
♦ loaded in memory protein sequence
♦ loaded in memory PB-fragments
♦ loaded HNCO, CBCACONH, and HNCA peak lists.
♦ specified tolerances.
Assignment>Contacts>HNCA
:: Calculate fragments contact map CHNCA
Assignment>Contacts>NOE>fawn
Results:
Assignment>Contacts>NOE>abacus
Assignment>Calculate Probabilities>SA
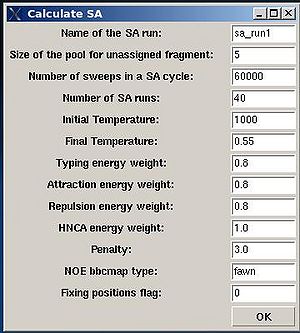
- “Name of the SA run”. Normally the name is sa_run#. A new directory under this name will be created within PROJECTNAME/assign directory. SA calculations will be curried out and the results will be stored in this directory.
- “Size of the pool for unassigned fragments”. The number of positions that are appended to the protein sequence and discarded (unassigned) fragments, if there are any, will be located there. It is safe to over-estimate this number. (If this number is under-estimated, this will force the mapping of spurious spin-systems onto protein sequence);
- “Number of SA trajectories”. The time needed for calculations is proportional to this number. On the other hand, having more SA trajectories the assignment probabilities could be calculated more accurately. In the case of good data, when all SA trajectories converge to assignments with the same energy, 10-15 trajectories should be enough. In the case of poor data, it is better to calculate 40-50 SA trajectories.
- “NOE bbcmap type”. User should specify which one NOE contact map, (abacus) or (fawn) should be used in the calculations;
- “Fixing position flag”. If the flag is set to 1, sequence position of all fragments which has assignment ID > -99 will be fixed during the simulation.
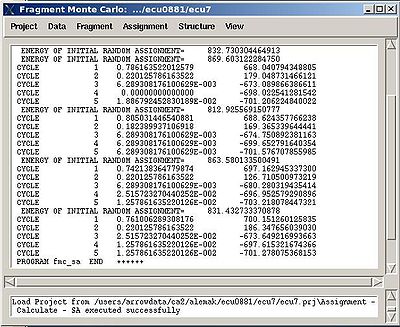
- “Final temperature”. Setting the optimal final temperature will provide all SA trajectories converge to optimal or sub-optimal assignments (the assignments that are in the vicinity of the global energy minimum). The optimal final temperature could be find by running one or a few SA runs with 3-4 trajectories and by analysing convergence of the trajectories from the report shown in the main FMCGUI window after each run (see Figure 2.10).
Assignment>Calculate Probabilities>REM
- “Name of the REM run”. Normally the name is rem_run#. A new directory under this name will be created within PROJECTNAME/assign directory. REM calculations will be curried out and the results will be stored in this directory.
- “Size of the pool for unassigned fragments”. The number of positions that are appended to the protein sequence and discarded (unassigned) fragments, if there are any, will be located there. It is safe to over-estimate this number. (If this number is under-estimated, this will force the mapping of spurious spin-systems onto protein sequence);
- “Number of REM steps”. The time needed for calculations is proportional to this number. On the other hand, with more REM steps more extensive sampling of assignment space wil be achieved, which in turn results in more accurate assignment probabilities. This number should be increased for large proteins.
- “NOE bbcmap type”. User should specify which one NOE contact map, (abacus) or (fawn) should be used in the calculations;
- “Fixing position flag”. If the flag is set to 1, sequence position of all fragments which has assignment ID > -99 will be fixed during the simulation.
- “Low temperature”. The optimal low temperature will provide extensive sampling of should sub-optimal assignments during REM simulation.
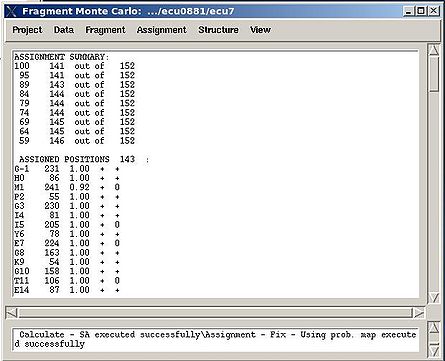
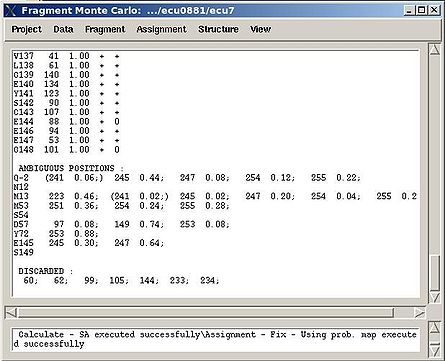
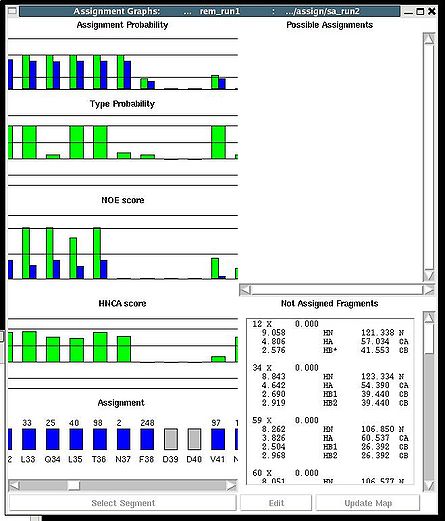
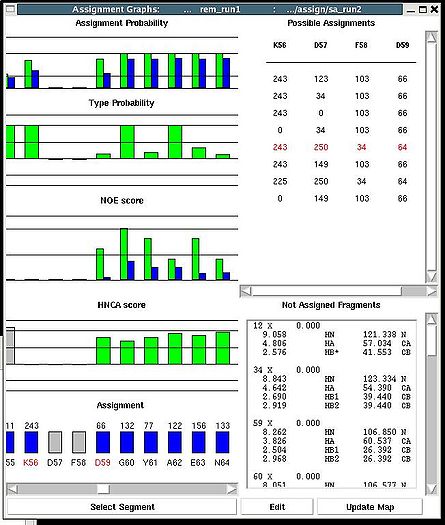
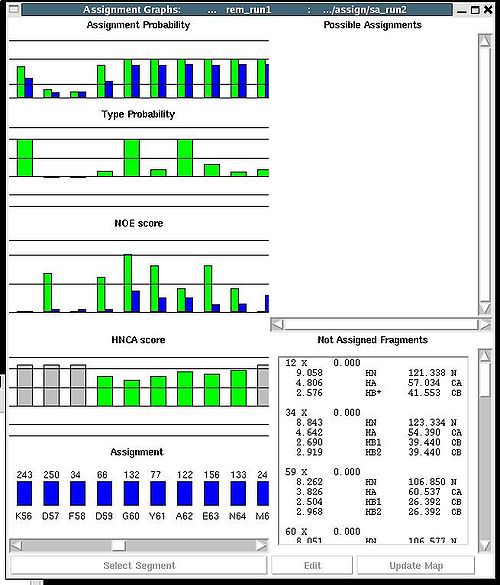
Assignment>Fix Assignment>Using Probability map
Results:
Assignment>Fix Assignment>Manually
♦ Assignment ID for selectedf PB fragments is specified
Assignment>Fix Assignment>Reset all
Assignment>Load Probabilities
Structure>Constraints>Talos>calculate
Structure>Constraints>Talos>load
Structure>Constraints>H-bonds>Specify
Structure>Constraints>H-Bonds>load
Structure>Calculate>Cyana
Structure>Calculate>ABCUS
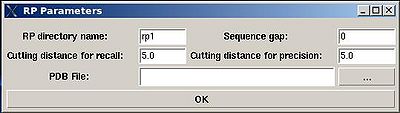
Structure>RPF>RP
Results:
♦ summary report
- “RP directory name”. Normally the name is rp#. A new directory under this name will be created within PROJECTNAME/assign directory. The results of the RPFanalysis will be stored in this directory.
- “sequence gap”. Residue pairs separated by less than the value of sequence gap will be excluded from generating expected peak lists.
- “cutting distance for recall”. Distance threshold for evaluating matching of an experimental peak to a structural ensemble (Recall score)
- “cutting distance for precition”. Distance threshold for generating expected peak from structural ensemble (Precision score)
Structure>RPF>DP
- : Set up calculations of DP score with AutoStructure
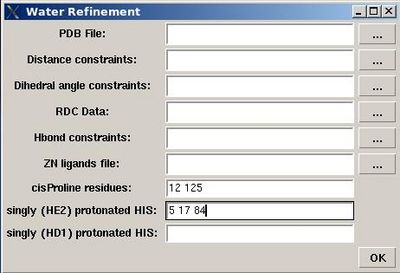
Structure>Water refinement>calculate
- specify the name of directory for water refinement calculations, WATDIR;
- select a number of files with coordinates and constraints;
- indicate cisProline residues (if there are any)
- specify protonation state of HIS residues (which is double protonated by default).
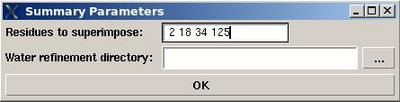
Structure>Water refinement>summary
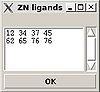
Structure>Add ZN ligands
Structure>RCI
:: Calculate Random Coil Index
View>Fragment
View>Assignment
:: Display and analize assignment results