XEASY Spin system identification: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 47: | Line 47: | ||
==== '''Analysis of the (3,2)D GFT HNNCO spectrum''' ==== | ==== '''Analysis of the (3,2)D GFT HNNCO spectrum''' ==== | ||
*Go to <tt>/protName/analysis/xeasy/hnco</tt> | |||
*In UBNMR run macro <tt>make32DHncoPeak:</tt> listed as following to get (3,2)D GFT HNNCO peaklist.<br> | |||
<blockquote>< | <blockquote><pre> | ||
read seq nhsqc.seq | init | ||
read prot noe.prot | read seq nhsqc.seq | ||
write prot hnco.prot | read prot noe.prot | ||
write prot hnco.prot | |||
simulate 2D N+pC HN | simulate 2D N+pC HN | ||
simulate 2D N-pC HN | simulate 2D N-pC HN | ||
write peaks HNNCO.peaks</ | write peaks HNNCO.peaks | ||
</pre></blockquote> | |||
*Do the analysis in XEASY on the (3,2)D GFT HNNCO spectrum similar to that of 3D HNNCO spectrum. | |||
<br> -- Main.GaohuaLiu - 16 Feb 2007</pre> <br> | <br> -- Main.GaohuaLiu - 16 Feb 2007</pre> <br> | ||
Revision as of 19:06, 10 November 2009
Spin System Identification
2D [15N,1H]-HSQC provide pairs of correlated amide 15N / 1HN chemical shifts. They seed spin systems - spins of individual residues, whose assignment to the protein sequence is normally not known a priori. In Xeasy-based GFT approach spin systems are called SRD spin systems.
(3,2)D HNNCO or 3D HNNCO provide additional resolution when both 15N and 1HN chemical shifts overlap, and help exclude side-chain peaks. Although 13C' chemical shifts are seldom used for sequence-specific assignment, they are used by CSI and TALOS programs.
Spin System Identification with XEASY/UBNMR
Peak Picking the (15N,1H) HSQC Spectrum
- Go to /protName/analysis/xeasy/nhsqc. Create a file with the amino acids sequence in FASTA format, obtained, for example, from the SPINE web page, and save it as aa.seq. In UBNMR, run makeSeq. This generates an XEASY <nop>SequenceList as nhsqc.seq . This <nop>SequenceList contains entries for the amino acids residues first, followed by two sets of SRDs named in the following SRD-I (Starting from 201) and SRD-II (starting from 401). SRD-II entires serve to handle sequential connectivities. See XEASY Files for Book-keeping: <nop>AtomList, <nop>SequenceList, <nop>PeakList.
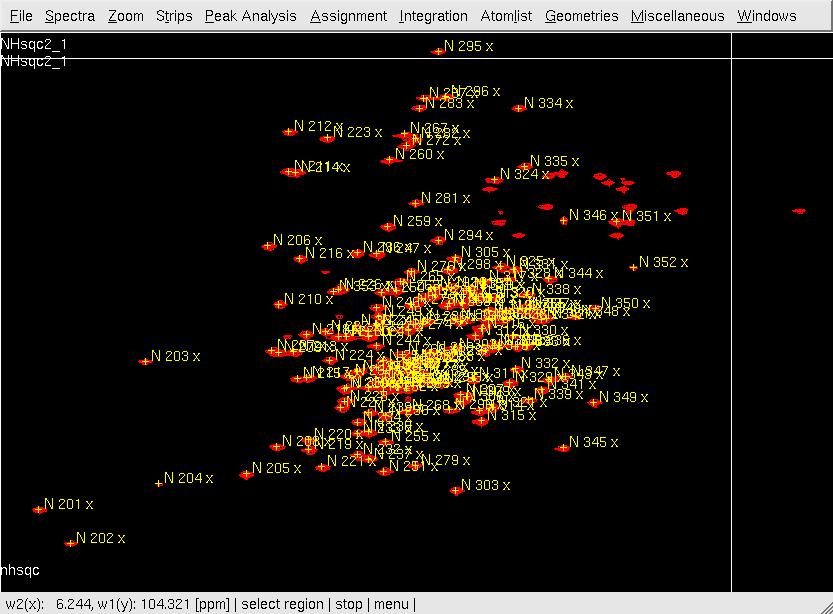
- In XEASY, use ns, ll and ls to load 2D [15N,1H]-HSQC spectrum, XEASY library and <nop>SequenceList. To change display from the default one to contour plot, type cp and give the appropriate threshold level. A corresponding initial <nop>AtomList is generated automatically; use in for in-phase 'peak picking' of the 2D [15N,1H]-HSQC spectrum automatically; complete peak picking manually by using dp to remove the peaks belonging to sidechain amides which can be identified by a NH2only HSQC if the region is crowded and pp to 'peak pick' additional peaks (Figure.1A); Use ar to automatically assign each peak to the backbone amide moiety of SRD-I residues (Figure.1B,C); use ac, wc and wp to save the updated <nop>AtomList as nhsqcO1.prot and <nop>PeakList as nhsqcO1.peaks. Then, 15N / amide 1HN chemical shifts are transferred into the <nop>AtomList entries corresponding to SRD-I. At times the ar command gives an error "Atom N 201 not known!". This can be rectified by loading a different library file [ll] from /nsm/chem/cen2/HTP2/3_src/xeasy/src.new/xeasy.lib and then trying to auto assign the peaks.
Figure 1. Peak picking the NHSQC spectrum*
A: After in-phase peak picking;
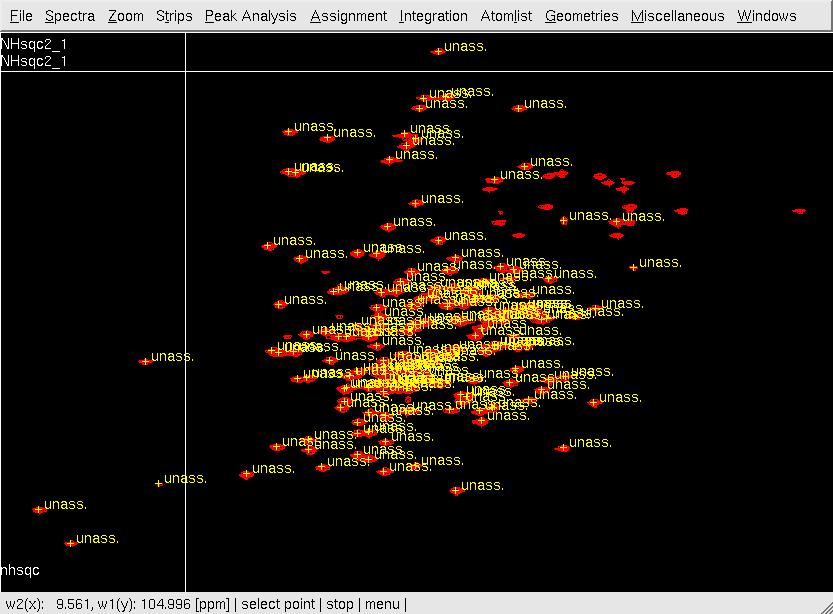
B: XEASY ar window
C: After XEASY command ar .
- In UBNMR run makeHncoPeak. A starting peak list for analysis of (3,2)D HNNCO or 3D HNNCO is generated as hncoI1.peaks. /
- In XEASY, perform HNNCO Analysis (described in HNNCO analysis).
Analysis of the 3D HNNCO Spectrum
- Go to /protName/analysis/xeasy/hnco
- In the spectra folder, make a identical copy of HNCO spectrum, eg. cp spectrum HNCO1 to spectrum HNCO1a.
- In UBNMR, run makeHncoPeak to produce the file hncoI1.peaks. This <nop>PeakList will contain one peak for each amide N,H moiety assigned to SRD-I derived from the 2D [15N,1H]-HSQC <nop>PeakList. 175 ppm is assigned to all 13C' shifts of SRD-II.
- In XEASY, use ns to load both HNNCO spectra HNCO1 and HNCO1a with permutation x:HN, y:C, Z:N and x:N, y:C, Z:HN, respectively. from /protName/xeasy/data; use ls to load the sequence file, nhsqc.seq; use lc to load the atom list (protlist), nhsqcO1.prot; use lp to load the initial HNCO peaklist from hncoI1.peak; use cp to display the spectrum as a contour plot; use se to create strips for all of the peaks in the peaklist; use gs to display the first set of strips (Figure 2A).
Figure 2: Analysis of the 3D HNNCO by XEASY
A: Before mr, showing orthogonal views of each strip;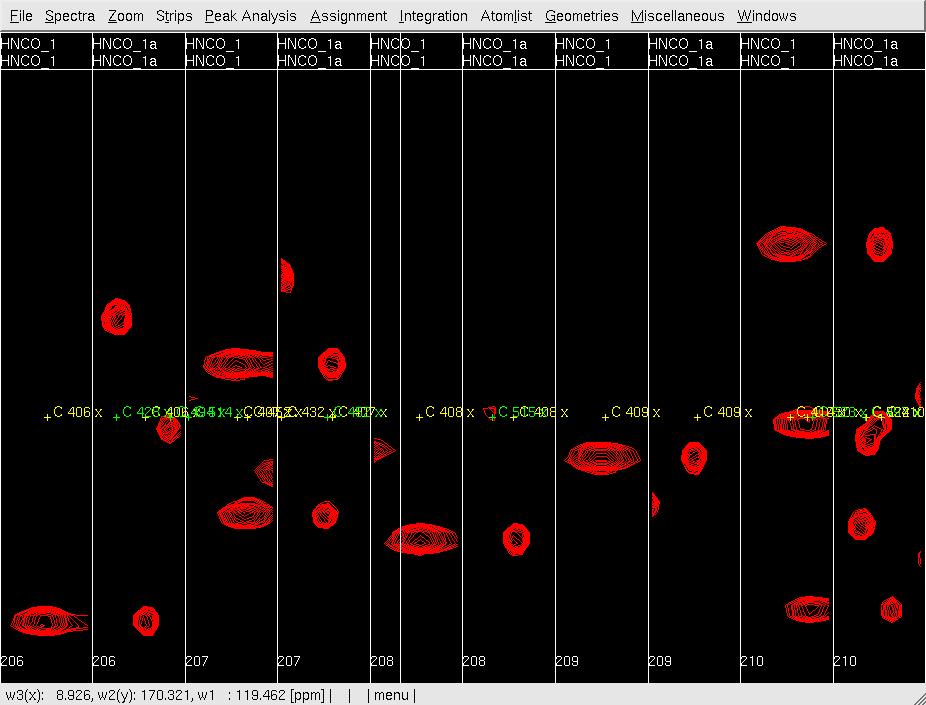
- In XEASY, use mr to move each pre-positioned peak onto the actual peak (Figure 2B); use dp to delete unobserved predicted peaks (side-chain peaks), and use pp manually pick and assign observed unpredicted peaks (overlapped in Nhsqc).
B: After mr;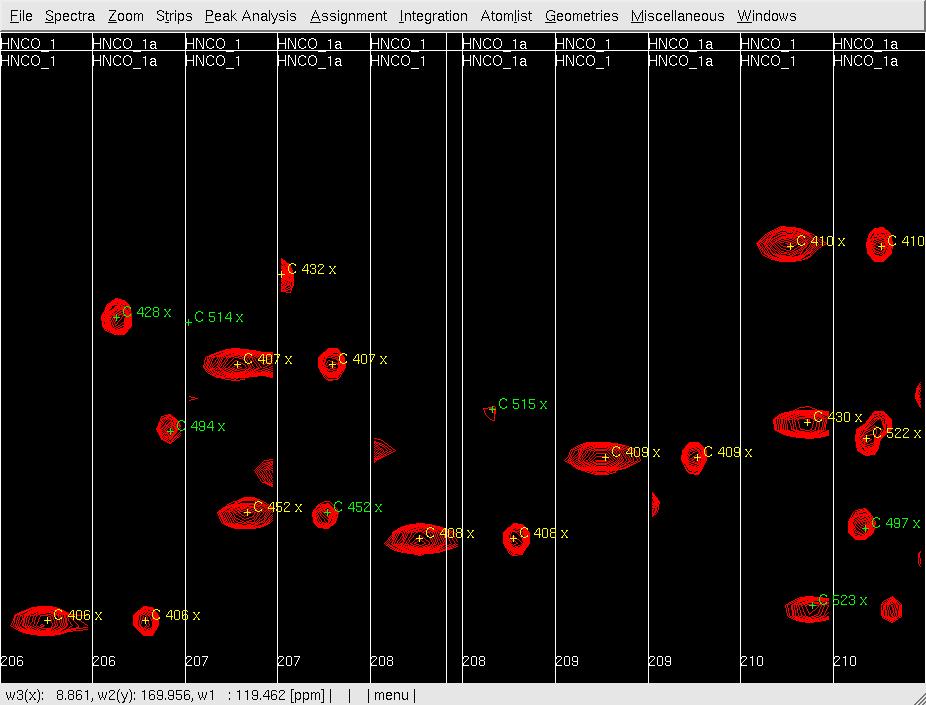
- After all peaks are 'picked', use ac to update the chemical shifts, wc to save the atom list as hncoO1.prot, and wp to save the peaklist as hncoO1.peaks.
- Go to next step for backbone assignment
Analysis of the (3,2)D GFT HNNCO spectrum
- Go to /protName/analysis/xeasy/hnco
- In UBNMR run macro make32DHncoPeak: listed as following to get (3,2)D GFT HNNCO peaklist.
init read seq nhsqc.seq read prot noe.prot write prot hnco.prot simulate 2D N+pC HN simulate 2D N-pC HN write peaks HNNCO.peaks
- Do the analysis in XEASY on the (3,2)D GFT HNNCO spectrum similar to that of 3D HNNCO spectrum.
-- Main.GaohuaLiu - 16 Feb 2007</pre>