Resonance Assignment/Abacus/FMCGUI objects: Difference between revisions
No edit summary |
No edit summary |
||
| Line 65: | Line 65: | ||
=== Contact map === | === Contact map === | ||
:There are three fragment contact maps: C<sup>f1</sup><sub> | :There are three fragment contact maps: C<sup>f1</sup><sub>HNCA</sub><span>(f2) , </span>C<sup>f1</sup><sub>NOE_B</sub><span>(f2) ,</span><span> and </span>C<sup>f1</sup><sub>NOE_F</sub><span>(f2) </span><span>, respectively. Each contact map scores the possibility for any fragment ''f1'' to be next to the fragment ''f1'' in protein sequence. Here ''f''1 and ''f2'' stand for fragment's user ID. </span> | ||
<div><span> Fragment contact map </span>C<sup>f1</sup><sub> | <div><span> Fragment contact map </span>C<sup>f1</sup><sub>HNCA</sub><span>(f2) </span><span>is calculated based on HNCA spectrum by the command {[[FMCGUI commands#Assignment.3EContacts.3EHNCA|Assignment>Contacts>HNCA]]}<div> Fragment<span> contact maps </span>C<sup>f1</sup><sub>NOE_B</sub><span>(f2) </span><span> and </span>C<sup>f1</sup><sub>NOE_F</sub><span>(f2) </span><span /><span> are calculated from NOESY spectra with or without using BACUS procedure, respectively.</span></div><div><span> </span><span> </span>C<sup>f1</sup><sub>NOE_F</sub><span>(f2) </span><span> can be calculated for all values of f1 and f2 by both commands {[[FMCGUI commands#Assignment.3EContacts.3ENOE.3Efawn|Assignment>Contacts>NOE>fawn]]} and {[[FMCGUI commands#Assignment.3EContacts.3ENOE.3Eabacus|Assignment>Contact>NOE>abacus]]}, while </span>C<sup>f1</sup><sub>NOE_B</sub><span>(f2) </span><span /><span> is calculated by command {[[FMCGUI commands#Assignment.3EContacts.3ENOE.3Eabacus|Assignment>Contact>NOE>abacus]]}.<div> </div></span></div></span></div> | ||
=== Assignment probabilities === | === Assignment probabilities === | ||
Revision as of 16:11, 1 December 2009
- protein sequence
- peak list
- and PB fragments.
Protein sequence
Peak list
- (+) peak list is required to be referenced
- (+/-) peak lists could be referenced optionally
- (-) peak lists for which referencing is not used even if it is present in the input file
N15 NOESY
|
-
|
C13 NOESY H2O
|
-
|
Arom NOESY
|
-
|
N15 HSQC
|
+
|
C13 HSQC
|
+
|
HNCA
|
+/-
|
HNCO
|
-
|
CBCACONH
|
+/-
|
HBHACONH
|
+
|
List of PB fragments
User ID
Fragment ID assigned by user, U_id. U_id can’t be changed within FMCGUI.
Assignment ID
Assignment ID, A_id, indicates the sequence position ID to which the fragment is assigned.
A_id = -99 if the fragment is not assigned to any position in the sequence.
A_id could be set up or modified by the following commands {Assignment>Fix Assignment>Manually}, {Assignment>Fix Assignment>Using probability Map}, and {Assignment>Fix Assignment>Reset all }.
Typing probabilities
Tt (f) is a probability for fragment f to have amino acid type t. Here t corresponds to one of 20 amino acid residue types, and f is fragment user ID.
Typing probbilities could be calculated or modified manually by the commands {Fragment>Type>Calculate} and {Fragment>Type>Fix}, respectively.
Contact map
- There are three fragment contact maps: Cf1HNCA(f2) , Cf1NOE_B(f2) , and Cf1NOE_F(f2) , respectively. Each contact map scores the possibility for any fragment f1 to be next to the fragment f1 in protein sequence. Here f1 and f2 stand for fragment's user ID.
Assignment probabilities
Ps(f) is a probability of fragment f to be assigned to sequense position s, where f is fragment user ID, and s is sequence position ID. There are two assignment probabilities associated with a fragment - PsSA(f) and PsREM(f) - that are calculated using Simulates Annealing (SA) and Replica Exchange Method (REM) Monte Carlo simulations, respistively.
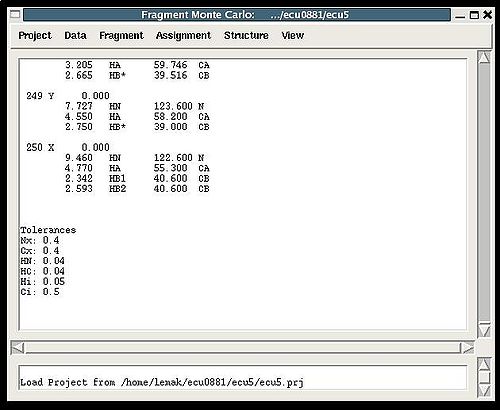
Main window
Figure 2.1
- the title bar displays the name of the current project and the directory inside which the project is located;
- the bar with six menus: Project, Data, Fragment, Assignment, Structure, and View, respectively;
- the project main window, where all messages from the last executed command are displayed ;
- the log window, where the history of executed commands is shown.