Resonance Assignment/CARA: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
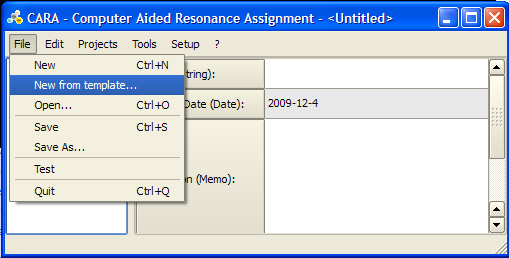
== Loading a New Template == | |||
To start a new structure determination project in CARA you need to load a template. A template is a CARA repository without project that contains definitions for residue types, spectrum type and LUA scripts. You can either load the default template from the [http://www.cara.ethz.ch/Wiki/TemplatesPage templates page] of the official CARA web-site, or extract a template from an existing CARA repository. | |||
For detailed instructions see: http://www.cara.ethz.ch/Wiki/ImportingTemplate | |||
<br>[[Image:CARA New Template Dialogue.png]] | |||
CARA Template for GFT spectra [[Media:BoR54_template.cara|BoR54_template.cara]]: Latest GFT template for BoR54 project | |||
Key modifications are: | |||
*Added linker residue definitions to facilitate treatment of homodimers | |||
*Added definitions of (4,3)D HNNCABCA, (4,3)D CABCA(CO)NHN and (4,3)D HABCAB(CO)NHN | |||
*Added attributes for GFT spectra - used by Lua scripts | |||
*LUA scripts | |||
**CreateConsensusPeaklist - consensus peaklist from two loaded peak lists | |||
**CreateSequentialSpinLinks | |||
**ExportSeqToAutostructure - prepare sequence in Autostructure format | |||
**ExportToAutoAssign - generate all files for AutoAssign input | |||
**ExportToCSI - generate CSI input file | |||
**GFT_aroHCCH_calc - small applet too calculate aromatic side-chain spin in (4,3)D HCCH-COSY | |||
**GFT_HCCH_calc - small applet too calculate aliphatic side-chain spin in (4,3)D HCCH-COSY | |||
**GFT_CABCA2CACB - calculate CA and CB from sum and difference shifts | |||
**ImportCyana2xPeakList - import peaklist in CYANA 2.x format (with multiple assignments) | |||
**ImportSparkyPeakList - import peaklist in Sparky format (from AutoStructure) | |||
**ImportFromAutoAssign - read assignments produced by AutoAssign | |||
**etc... | |||
#[[CARA Introduction|Introduction]] | #[[CARA Introduction|Introduction]] | ||
#[[Spin System Identification with CARA|Spin System Identification in 2D 15N-HSQC and 3D HNNCO]] | #[[Spin System Identification with CARA|Spin System Identification in 2D 15N-HSQC and 3D HNNCO]] | ||
Revision as of 23:09, 3 December 2009
Loading a New Template
To start a new structure determination project in CARA you need to load a template. A template is a CARA repository without project that contains definitions for residue types, spectrum type and LUA scripts. You can either load the default template from the templates page of the official CARA web-site, or extract a template from an existing CARA repository.
For detailed instructions see: http://www.cara.ethz.ch/Wiki/ImportingTemplate
CARA Template for GFT spectra BoR54_template.cara: Latest GFT template for BoR54 project
Key modifications are:
- Added linker residue definitions to facilitate treatment of homodimers
- Added definitions of (4,3)D HNNCABCA, (4,3)D CABCA(CO)NHN and (4,3)D HABCAB(CO)NHN
- Added attributes for GFT spectra - used by Lua scripts
- LUA scripts
- CreateConsensusPeaklist - consensus peaklist from two loaded peak lists
- CreateSequentialSpinLinks
- ExportSeqToAutostructure - prepare sequence in Autostructure format
- ExportToAutoAssign - generate all files for AutoAssign input
- ExportToCSI - generate CSI input file
- GFT_aroHCCH_calc - small applet too calculate aromatic side-chain spin in (4,3)D HCCH-COSY
- GFT_HCCH_calc - small applet too calculate aliphatic side-chain spin in (4,3)D HCCH-COSY
- GFT_CABCA2CACB - calculate CA and CB from sum and difference shifts
- ImportCyana2xPeakList - import peaklist in CYANA 2.x format (with multiple assignments)
- ImportSparkyPeakList - import peaklist in Sparky format (from AutoStructure)
- ImportFromAutoAssign - read assignments produced by AutoAssign
- etc...