Resonance Assignment/Abacus/Introduction to ABACUS: Difference between revisions
Jump to navigation
Jump to search
Figure 1.2
Figure 1.4
RyanDoherty (talk | contribs) No edit summary |
|||
| Line 1: | Line 1: | ||
= ABACUS approach. = | = ABACUS approach. = | ||
<div>ABACUS (''A''pplied ''BACUS'') is a novel approach for protein structure determination that has been applied successfully for more than 20 NESG targets. ABACUS is characterized by use of BACUS, a procedure for automated probabilistic interpretation of NOESY spectra in terms of unassigned proton chemical shifts based on the known information | <div>ABACUS (''A''pplied ''BACUS'') is a novel approach for protein structure determination that has been applied successfully for more than 20 NESG targets. ABACUS is characterized by use of BACUS, a procedure for automated probabilistic interpretation of NOESY spectra in terms of unassigned proton chemical shifts based on the known information about the "connectivity" between proton resonances. BACUS is used in both the resonance assignment and structure calculation steps. The resonance assignment strategy of ABACUS<span> is what distinguishes it the most from conventional NMR structure determination approaches (see Fig.1.1A). </span><br></div> | ||
==== '''Figure 1.1A''' ==== | ==== '''Figure 1.1A''' ==== | ||
[[Image:Abacus.JPG|thumb|left|350px]]<br> | [[Image:Abacus.JPG|thumb|left|350px]]<br> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> ''' ''' | <br>''' ''' | ||
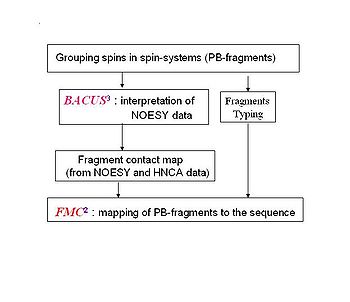
Flowchart of resonance assignmnent by ABACUS''. '' | Flowchart of resonance assignmnent by ABACUS''. '' | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | <br> | ||
<br> | |||
==== '''<span />''' ==== | |||
==== '''<span>Some features /advantages of the ABACUS protocol:</span>''' ==== | ==== '''<span>Some features /advantages of the ABACUS protocol:</span>''' ==== | ||
| Line 36: | Line 38: | ||
*It can make use of partial spin-systems; | *It can make use of partial spin-systems; | ||
*It can efficiently identify manual errors in the input peak lists; | *It can efficiently identify manual errors in the input peak lists; | ||
<div></div> | <div></div> | ||
<br> | <br> | ||
| | ||
Revision as of 18:46, 6 January 2010
ABACUS approach.
ABACUS (Applied BACUS) is a novel approach for protein structure determination that has been applied successfully for more than 20 NESG targets. ABACUS is characterized by use of BACUS, a procedure for automated probabilistic interpretation of NOESY spectra in terms of unassigned proton chemical shifts based on the known information about the "connectivity" between proton resonances. BACUS is used in both the resonance assignment and structure calculation steps. The resonance assignment strategy of ABACUS is what distinguishes it the most from conventional NMR structure determination approaches (see Fig.1.1A).
Figure 1.1A
Flowchart of resonance assignmnent by ABACUS.
Some features /advantages of the ABACUS protocol:
- It does not rely on sequential connectivities from less sensitive experiments such as HNCACB indispensable for most traditional sequential assignment procedures;
- Inter-residue sequential connectivities are established mainly from NOE data, which saves time at a later stage in “troubleshooting” NOE and resonance assignments.;
- Probabilistic nature of the ABACUS procedure provides measure of reliability of assignments, and therefore one can obtain a partial, yet highly reliable assignment (even when the NMR data are sub-optimal) with the knowledge of where to focus manual intervention;
- It can make use of partial spin-systems;
- It can efficiently identify manual errors in the input peak lists;
NMR spectra required for ABACUS
The spectra typically needed for ABACUS approach are most conveniently separated into 3 groups: NH-rooted, the CH-rooted and the aromatic (also CH-rooted). Table 1 shows the optimal set of NMR spectra. This, of course, is neither an exclusive or exhaustive list. For example, a simultaneous CN-NOESY could be recorded instead of three different ones listed in the table. In case there are very few aromatic residues in a protein, to collect only one aromatic spectrum, namely aromatic NOESY, could be enough for assignment of aromatic resonances.
Table 1. ABACUS optimal set of experiments
NH-rooted
|
CH-rooted
|
Aromatic
|
15N-HSQC
|
13C-CT-HSQC
|
13C-HSQC-aro
|
HNCO
|
13C-HSQC
|
H(C)CH-TOCSY-aro
|
HNCA
|
H(C)CH-TOCSY
|
(H)CCH-TOCSY-aro
|
CBCA(CO)NH
|
(H)CCH-TOCSY
|
13C-NOESY-HSQC-aro
|
HBHA(CO)NH
|
13C-NOESY-HSQC
|
|
15N-NOESY-HSQC
|
||
CCCONH-TOCSY (optional)
| ||
H(CCCO)NH-TOCSY (optional)
| ||
Spin-system identification strategy
The resonance assignment procedure starts from grouping resonances in spin systems. Two kind of spin-system will be considered in this manual.
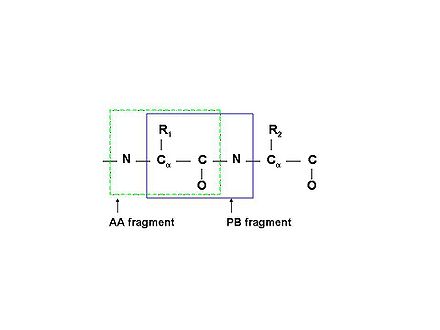
PB fragment
PB (Peptide Bond) fragment comprise correlated resonances from the side chain of residuei and the NH resonances of residue i+1 (see Figure 1.1B).
Figure 1.1B
Schematic description of two types of molecular fragments: traditional spin-system (AA-fragment)include all the atoms belonging to the same residue; PB-fragment includes all the atoms from one residue except the backbone amide group, plus the amide group from the next residue in the protein
bPB fragment
Uncompleted HN-rooted PB spin-systems, which include resonances of Cα, Hα, Cβ, and Hβ atoms of residue i and the NH resonances of residue i+1, is called bPB fragment.
Spin-system identification in ABACUS approach consists of 3 main steps.
1. On the first step, bPB fragments are collected from high sensitivity NMR correlation experiments (such as HNCO, CBCA(CO)NH, and HBHA(CO)NH) that transfer magnetization via the intervening peptide bond (see Figure 4.1A).
2. On the second step, completion of bPB fragments with side-chain aliphatic resonances as well as identification of additional spin-systems (lacking HN resonances) is performed using HCCH-TOCSY and 13C-NOESY spectra (see Figure 4.1B)
3. Finally, spin-system validation and correction is performed. This step allows one to find mistakes made during spectra peak-picking and to correct the mistakes by going back to the spectra.
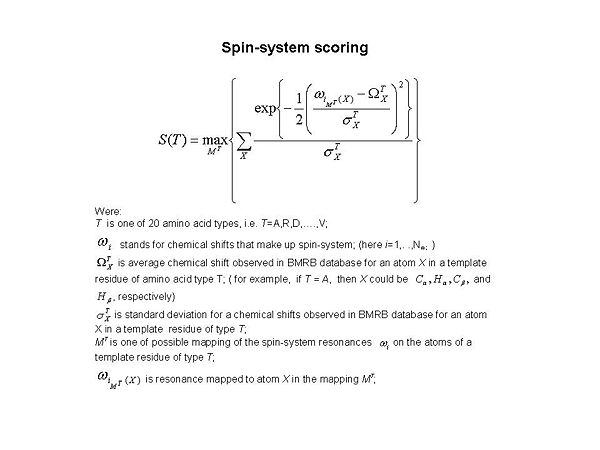
Figure 1.2
For each spin-system, 20 scores S(T) were calculated during the validation (see Figure 1.2). Here T corresponds to amino acid type, and T=A,R,D,…, and V, respectively. The score evaluate goodness-of-fit of the spin-system resonances to those observed in BMRB data base. Too low value of the best score Smax, where Smax = max{ S(T)} , means that either the spin-system has very unusual chemical shifts or the spin-system does not make sense and need to be
corrected.
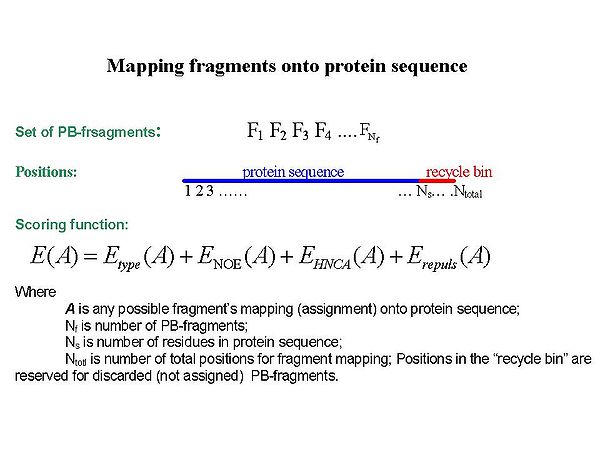
Fragment assignment by FMC procedure
Sequence-specific assignment of PB-fragments is achieved using a Fragment Monte Carlo (FMC) stochastic search procedure. The scoring function used in the FMC procedure is based on both fragment amino acid typing (matching the spin system to amino acid types) and fragment contact map (reflecting which residue is next to which) derived from HNCA data and the analysis of NOEs interpreted by BACUS (see Figure 1.3)
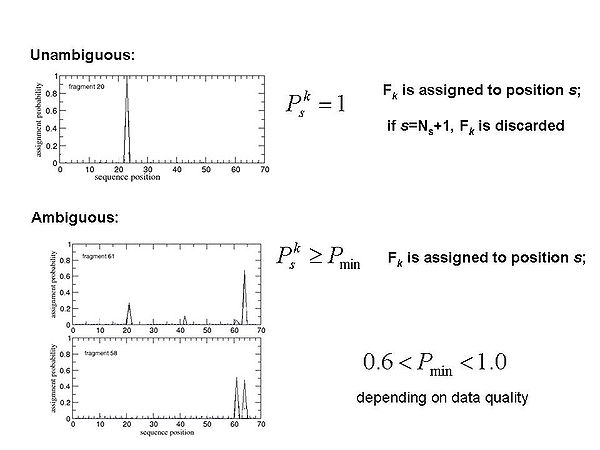
Figure 1.3
FMC procedure performs probabilistic assignment of PB-fragments. The assignment probabilities Psk are calculated by Simulated Annealing (SA) or Replica Exchange Method (REM) Monte Carlo (MC) simulations. Here, Psk is a probability of fragment k to occupy position s;'k = 1,….,Nf. ;
Figure 1.4
FMC Graphical User Interface
FMCGUI is a graphical interface that assist user to carry out resonance assignment and structure calculation using ABACUS approach. FMCGUI integrate a number of FORTRAN applications: performs control of the data-flow between the applications, execute the applications, and helps to analyze effectively obtained results by visualizing data.
The main purpose of FMCGUI is to provide interactive tool for resonance assignment.
Structural part of FMCGUI can be used independently from the resonance assignment part. It helps to set up both structure calculations with CAYNA and water refinement calculations with CNS and to analyse results. The actual structure calculations are supposed to be carried out outside FMCGUI on linux cluster.