Resonance Assignment/AutoAssign
Introduction
AutoAssign is a constraint-based expert system for automating the assignment and analysis of backbone NMR resonance assignments of proteins (Ref. 1). The program is implemented in C++, Java2, and Perl programming languages and supported on SGI-IRIX, Sun-Solaris, MAC-OSX, x86-Linux, and x86_64-Linux architectures. The newest AutoAssign distribution (version 2.4.0) automates the assignments of HN, NH, CO, CA, CB, HA, and HB resonances in non-, partially-, and fully-deuterated samples. The rich graphical user interface (GUI) provides a many sets of tools for dataset conversions, assignment validations, and various graphical displays of assignment results. AutoAssign is well tested on a large number of independently-collected triple-resonance NMR data sets of proteins ranging in size from ~6 to ~32 kD, including one fully-deuterated protein and and a dataset with reduced-dimensionality experiments. AutoAssign performs the automated analysis of assignments in only seconds on current RISC and x86 platforms.
The following description is mainly taken from the AutoAssign SOP (tutorial). Please check this tutorial and the version 2.4.0 help documentation for more information and usage.
Using AutoAssign
Input Files
The input files for AutoAssign are:
- A sequence file in fasta format
- Peak lists for backbone experiments in Sparky format.
Opening AutoAssign
- Run the startup script
autoassignIf you are running the client on the same machine as the server. - If you are running the client on a machine different from the server, then you must first make sure that the server is running on the other machine. The easiest way to do this is to run the startup script
autoserveron this other computer.
The main AutoAssign window will appear (Figure 1).
Figure 1: AutoAssign Main Window
Creating a Data Set
Using the Peaklist Converter in the Graphical User Interface
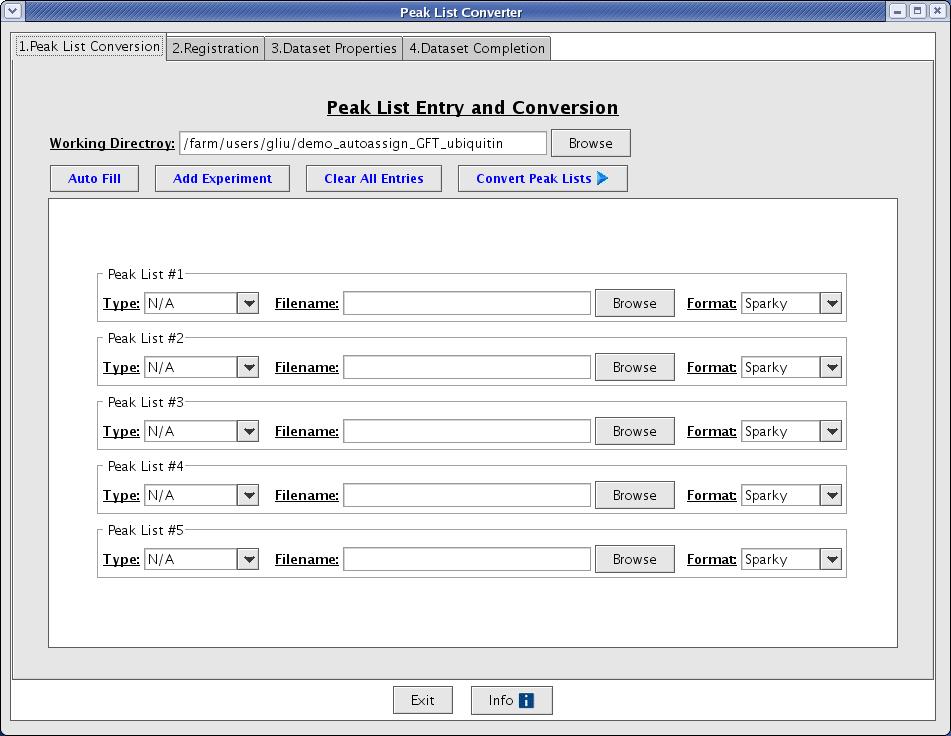
The easiest way to create a dataset is to using the Dataset Creation Graphical User Interface. This interface will guide you through the basic steps of converting your peak lists, registering your peak lists, creating a control file, and evaluating the quality of your peak lists. Go to File -> Convert/Create Dataset from the main menu option to start the Dataset Creation Graphical User Interface (Figure 2).
Figure 2: The Peak List Converter
The process is divided into 4 steps: 1. Peak list conversion, 2. Registration, 3. Data set properties, and 4. Data set completion.
Peak list conversion
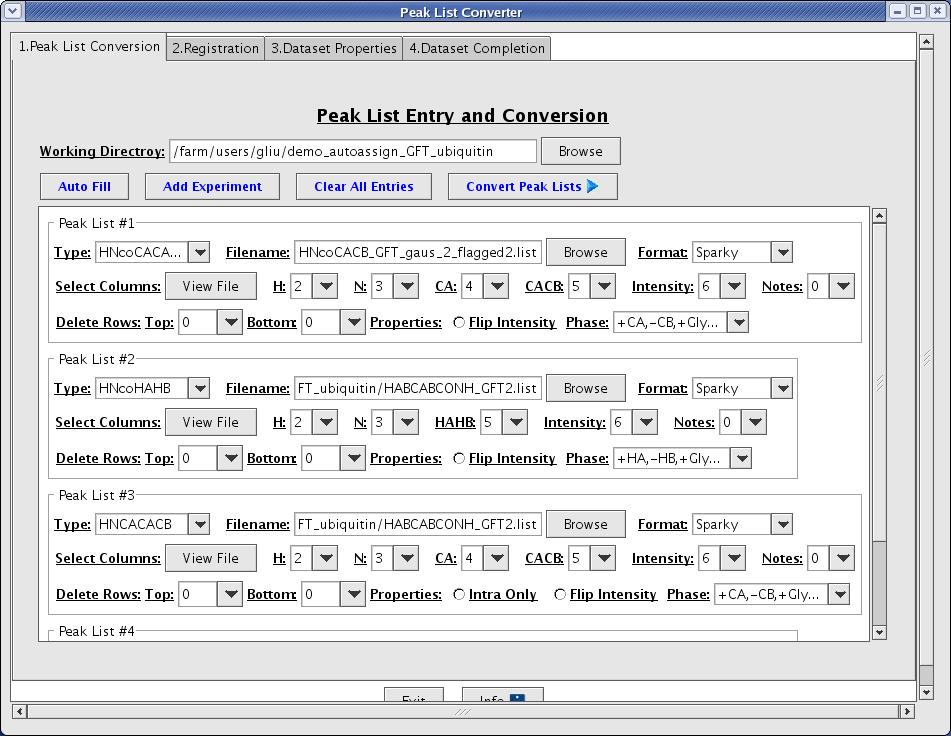
The peak list conversion process is designed to reformat given peak lists into AutoAssign format (Figure 3). Here the user defines the columns and spectral properties of each peak list.
Figure 3: Peak List Conversion Page
- Working directory [text field]: it is a base directory that the interface will use to create a subdirectory with a time stamp. The default is set to the current working directory.
- Auto Fill [button]: when a user decides to reuse the same peak lists or the same type of configuration based on the previous analysis, this function allows a user to import the data previously used.
- Add Experiment [button]: This function allows a user to display additional blank peaklist panel.
- Clear All Entries [button]: this function clears populated data fields displayed on screens.
- Convert Peak Lists [button]: this function runs a set of AutoAssign perl scripts to converts peak lists entered by a user. It generates intermediate files used to compute registration/tolerance values for the subsequent steps. All scripts executions run by interface is logged in
event.logfile found under the subdirectory named with a time stamp along with other intermediate files.
Registration
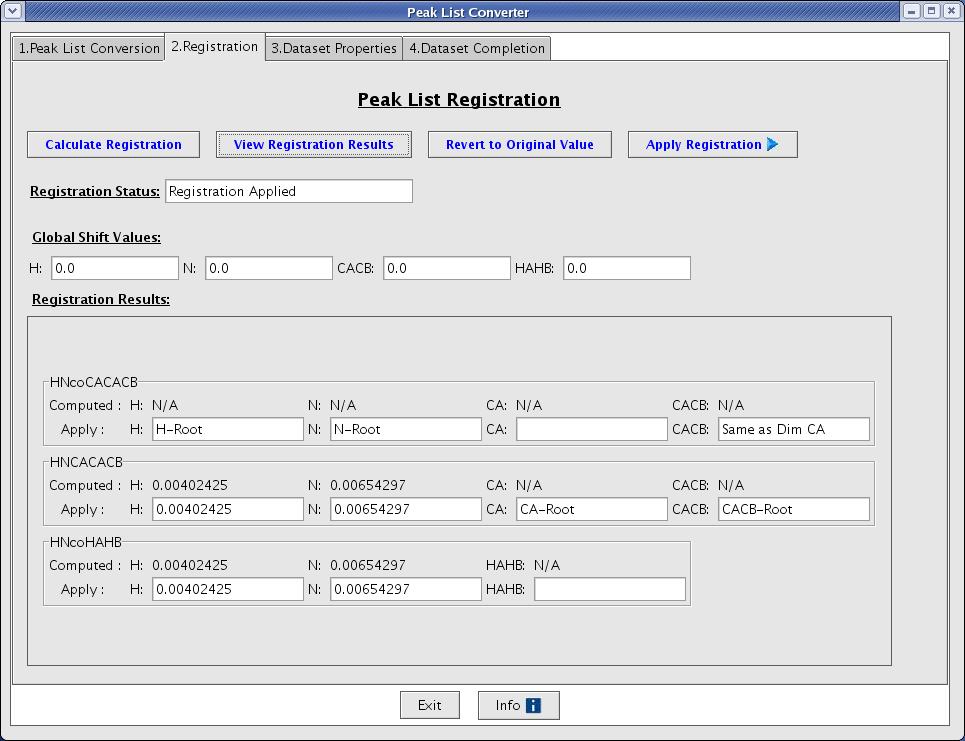
A user will be taken to the Registration tab when Convert Peak List button is clicked. This will allow a user to calculate registration values and apply those values to shift peaks by creating new / temporal peaklist files (Figure 4).
Figure 4: Peak List Registration Page
- Calculate Registration [button]: this function starts calculating registration value and it will open a progress monitor window to notify you how many calculations are completed. The error may be recorded in .reg file(s) under subdirectory if it occurs.
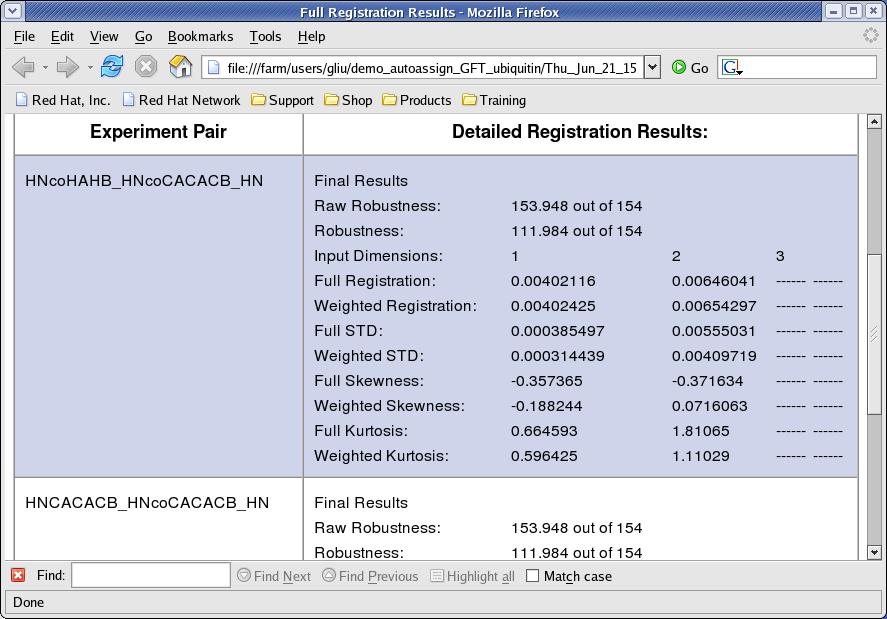
- View Registration [button]: the interface will create an html page by consolidating .reg files generated and will try to open it with a browser. The
Weighted Registrationvalues are used to register peak lists. TheFull Stdvalues are used in calculating tolerances included in the control file (Figure 5). - Revert to Original Value [button]: this function refills text fields for registration value with original registration value calculated.
- Apply Registration [button]: this function applies registration values to each peaklist. It creates new peaklist files with _shifted.pks extension.
Figure 5: Registration Results Page
References
1. Zimmerman, D.E., Kulikowski, C.A., Huang, Y., Feng, W., Tashiro, M., Shimotakahara, S., Chien, C., Powers, R. and Montelione, G.T. (1997) Automated analysis of protein NMR assignments using methods from artificial intelligence. J. Mol. Biol. 269, 592-610.