AGNuS/AutoProc
Introduction
Autoproc/AGNuS is a suite of programs for the automatic generation of scripts for multidimensional protein NMR data processing and format conversion. AGNuS automatically generates NMRPipe scripts for processing multidimensional Varian or Bruker NMR experiments and provides functions for chemical referencing. AGNuS calls information for specific experiments and spectrometers stored in table files, which can be generated/modified by users with permission. AGNuS is also capable of processing reduced dimensionality/GFT experiments.
The current version of the program is AutoProc 2.0. Please check the documentation and examples provided with the software for addtional information.
How to Use AGNuS
Processing a 2D 1H,15N-HSQC Using AGNuS
AGNuS parses procpar (Varian) and acqu* (Bruker) files for spectral parameters. The program consults a pulse sequence table specific to the sequence used in the experiement. For example, this is the table file for the standard Bruker 2D 1H,15N-HSQC, hsqcetf3gpsi. If the sequence table file is missing from the Pulse_Sequences directory in AGNuS, the program will through an error, and the user must create one (best to clone one from an similar one already in the directory).
# This is a Bruker experiment and only a table for testing. file_type : single_fid_table name : hsqcetf3gpsi nickname : HSQC dimensions : [ nuclei : H offset_parameter : O1_1 acquisition_mode : DQD ] dimensions : [ nuclei : N offset_parameter : O1_2 acquisition_mode : Echo-AntiEcho ] processing_defaults : [ name : "Sine Bell Default" suffix : _SP dimensions : [ transformation : FT apodization : "SP -off 0.45 -end 0.98 -pow 2" first_point_correction : 0.5 phase0 : -60 ] dimensions : [ transformation : FT apodization : "SP -off 0.45 -end 0.98 -pow 2" first_point_correction : 0.5 phase0 : -90 ] ]
Start autoproc and enter a spectrometer, then a DSS value for referencing. This is the most optimal way to reference the spectrum. We reference to internal DSS in all of our production work.
AGNuS 2.1 Type "agnus -help" for help Documentation is located at URL file:/farm/software/NewAutoProc2.0/help_files/index.html WARNING *** SW_h (2000) does not equal SW * SFO1 (4865.4880848234). Setting SW_h = SW * SFO1. Experiment hsqcetf3gpsi selected. ** Spectrometer Selection Menu ** 1) CABM_Bruker_500 2) CABM_Bruker_600_100306 3) CABM_Varian_500 4) CABM_Varian_500_060805_20C 5) CABM_Varian_600 6) NYSBC_800_Conventional 7) PNNL_Baker_600 8) PNNL_Glacier_600 9) PNNL_Varian_500 10) PNNL_Varian_750 11) PNNL_Varian_800 12) SUNY_Buffalo_Varian_600 13) SUNY_Buffalo_Varian_750 14) Z_500_No_Reference 15) Z_600_No_Reference 16) Z_750_No_Reference 17) Z_800_No_Reference Spectrometer [1]: 17 ** Conversion Parameter Selection Menu ** -- Current Directory: /farm/data/jma/JIM_NESG_PROJECTS/PfR193A_PROJECT/FIDs/B800/PfR193A.019_NC5_062509/NHhsqc_largeSW1_exp4 -- Input Directory: ./ 1) Input Data File (FID) Name: ser 2) Working Directory: ./ -- Conversion Subdirectory: pipe_conversion/ 3) NMRPipe Output Name: HSQC.fid 4) Conversion Script Directory Option: working_dir 5) Conversion Script Name: Convert_HSQC.csh 6) Experiment Temperature (celsius) (from procpar/acqus file): 25 7) Absolute Reference of DSS at experiment conditions (in MHz): Select parameter to change or [Return] to continue: 7 WARNING: This overrides default behavior of AGNUS. Default behavior uses the DSS temperature referencing coefficients in the spectrometer table file. Leave this field blank or zero if you do not wish to override standard behavior. Absolute Frequency of DSS: 800.1899230
This produces the following NMRPipe conversion file, which the user then can modify and subquently run:
Conversion script: ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ #!/bin/csh # This hsqcetf3gpsi conversion script was created using AGNuS 2.1. # Make the conversion directory if it does not exist (because var2pipe will not for 2D's) if (! -d ./pipe_conversion/ ) mkdir ./pipe_conversion/ bruk2pipe -bad 0.0 -noaswap -DMX -decim 16 -dspfvs 12 -in ./ser \ -xN 2048 -yN 256 \ -xT 1024 -yT 128 \ -xMODE DQD -yMODE Echo-AntiEcho \ -xSW 11160.7142857143 -ySW 4865.4880848234 \ -xOBS 800.19376489395 -yOBS 81.09146808039 \ -xCAR 4.80122760796774 -yCAR 110.121740240004 \ -xLAB 1H -yLAB 15N \ -ndim 2 -aq2D States \ -out ./pipe_conversion/HSQC.fid -verb -ov ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ Known Nuclei: H,N
A default NMRPipe processing script is then generated, which can again be modified by the user, and subequently run:
Full Processing script: ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ [1] #!/bin/csh [2] # [3] # This hsqcetf3gpsi processing script was created using AGNuS 2.1. [4] # [5] # 2D Processing Script. [6] [7] nmrPipe -in ./pipe_conversion/HSQC.fid \ [8] | nmrPipe -fn SOL \ [9] | nmrPipe -fn SP -off 0.45 -end 0.98 -pow 2 -c 0.5 \ [10] | nmrPipe -fn ZF -auto \ [11] | nmrPipe -fn FT \ [12] | nmrPipe -fn PS -p0 -60 -p1 0 -di \ [13] | nmrPipe -fn TP \ [14] | nmrPipe -fn SP -off 0.45 -end 0.98 -pow 2 -c 0.5 \ [15] | nmrPipe -fn ZF -auto \ [16] | nmrPipe -fn FT \ [17] | nmrPipe -fn PS -p0 -90 -p1 0 -di \ [18] -verb -ov -out ./HSQC_SP.pipe [19] ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
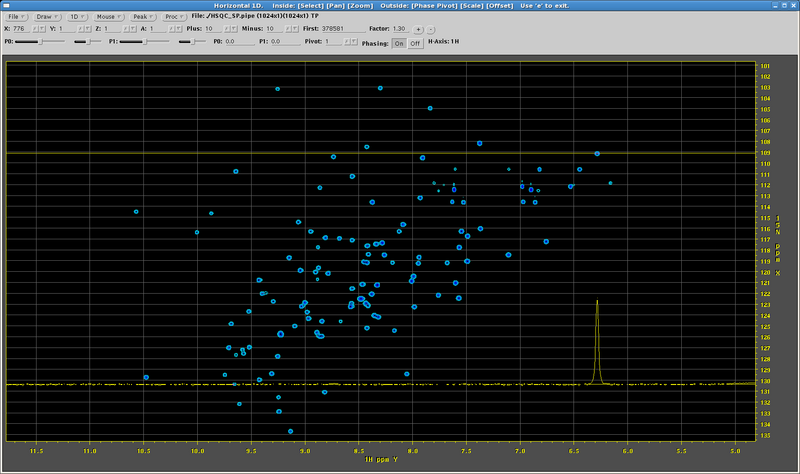
Next the program launches NMRDraw (Figure 1). Here the user determines phase corrections in both dimensions, which are then applied to the processing script and the data is reprocessed.
Figure 1: An NMRDraw Window for a 1H,15N-HSQC
Finally, the user converts to SPARKY or XEASY format, and proceeds to spectral analysis.
Processing a 3D Experiments Using AGNuS
Processing higher dimensionality experiments using AGNuS is essentially the same process with a few minor differences. For example, for a 3D experiment, the user first produces F1F3 (xy) and F2F3 (xz) 2D planes which are phase corrected using NMRDraw. Then the 3D NMRPipe script is run to produce the 3D processed pipe file.
Processing a GFT Experiment Using AGNuS
Here are some instructions for processing a GFT experiment:
- Before running AGNuS, an appropriate pulse sequence table corresponding to the NMR data should be prepared. The table files for the most used NMR experiments are availble from AGNuS. AGNuS will automatically detects the procpar file, parses it, and selects the appropriate parameters from the procpar and the corresponding experiment description (pulse sequence table). Select
*MULTIfor GFT basic experiments process.
Example of pulse sequence table for GFT_43D_CABCAcoNHN
file_type : single_fid_table
name : GFT_43D_CBCACAcoNHN
nickname : HNcoCACB_GFT
synonym : GFT_43D_CBCACAcoNHN_MULTI
dimensions : [
nuclei : H
offset_parameter : tof
acquisition_mode : Complex
]
dimensions : [
nuclei : C
offset_parameter : dof
acquisition_mode : Complex
]
dimensions : [
nuclei : N
offset_parameter : dof2
acquisition_mode : Complex
]
processing_defaults : [
name : "Sine Bell Default"
dimensions : [
transformation : FT
apodization : "SP -off 0.45 -end 0.98 -pow 2"
first_point_correction : 0.5
]
dimensions : [
transformation : FT
apodization : "SP -off 0.45 -end 0.98 -pow 1"
first_point_correction : 1.0
]
dimensions : [
transformation : "FT -neg"
apodization : "SP -off 0.45 -end 0.98 -pow 1"
first_point_correction : 0.5
]
xy_plane : 002
]
- To run AGNuS, simply go to the directory only contains the Varian fid and procpar (parameter) file; type
agnusand follow the instructions prompted by AGNuS. Typeagnus -helpfor help.- Select
*MULTIfor GFT basic experiments process (optional, AGNuS will go to the next step if non-GFT data if identified) - Selecting the spectrometer
- Select the basic information about where files are located, name of conversion scripts, and names of converted files.
- Run the conversion script provided by AGNuS to convert NMR data format to Pipe format. Modify the conversion script is necessary.
- Run the process scripts (3D and XY, XZ planes process scripts for 3D data) provided by AGNuS. Modify the Buffalo.NMRPipe process parameters such as phase values if necessary.
- AGNuS will lauch NMRDraw to let the user to check the process data and get the phase values.
- Use
CTRL-Cto quit AGNuS after phasing the spectrum with NMRDraw - Rerun AGNuS and manually type in or correct the phase when come to the process scripts change step. Modify the base line parameter as well if necessary
- Run the final process after all parameters be carefully reviewed.
- Run the UCSF conversion and cleanup script if use SPARKY for NMR data analysis. Run SPSCAN to conver Buffalo.NMRPipe format data to XEASY format data if use XEASY.
- Select
-- GaohuaLiu - 31 May 2007
-- Updated by JimAramini - 11 Nov 2009