Structure Refinement Using CNS Energy Minimization With Explicit Water
Introduction
Crystallography & NMR System (CNS) is a program has been designed to provide a flexible multi-level hierachical approach for the most commonly used algorithms in macromolecular structure determination [1]. Functions include heavy atom searching, experimental phasing (including MAD and MIR), density modification, crystallographic refinement with maximum likelihood targets, and NMR structure calculation using NOEs, J-coupling, chemical shift, and dipolar coupling data.
The latest version is 1.21. Please check the website http://cns-online.org for additional information.
CNS Refinement Protocol
The final structure from CYANA should be refined using CNS energy minimization with explicit water before PDB deposition.
The common input files are:
- PDB coordinates
- NOE constraints
- Dihedral angle constraints
- Hydrogen bond constraints
Converting input files from CYANA to XPLOR/CNS
If the final structure calculation was performed with CYANA 2.1, then the constraint and coordinate files must be first converted to XPLOR/CNS format.
Using cyana2cns.cya script
This procedure can be used to prepare input files to use with the WaterRefCNS script (see below).
- Download cyana2cns.cya into directory with the final structure calculation.
- Modify the read pdb, read upl and read aco lines if necessary.
- Start CYANA 2.1 and run cyana2cns.cya.
- All conformers will be stored in a single PDB file KKK.pdb. If you plan run water bath refinement manually, use the p2X program instead.
- The resulting NOE constraint file KKK_noe.tbl will have all lower limits set to 0. If want them to be set according to VdW radii, use the r2X program instead. Experience shows that there is no significant effect on geometry and quality scores as reported by PSVS.
Using programs p2X and r2X and d2X
These programs were written by Alex Lemak at the University of Toronto. They are typically used to prepare input files for manual CNS water bath refinement.
Go to ~1_projects/targetID/structure/cns/ directory and conversion_scripts sub directory, convert required input files to CNS format from CYANA format by using macro mkfil:
p2X cyana2.pdb pref
r2X found-c.upl noeupl.tbl
r2X gr-hbonds.upl hbonds.tbl
cat noeupl.tbl hbonds.tbl > noe.tbl
d2X found-c.aco aco.tbl
- p2X splits conformers from a pdb file generated by CYANA 2.1 into individual pdb files (one file per conformer) and converts atom names to CNS format. The second argument "pref" specifies a common prefix name for the output pdb files. It should be no more than 4 characters long.
- r2X converts distance constraints (noe/hbond) from CYANA 2.1 format to CNS format. The lower limits in the resulting constraint file are set according to VdW radii.
- Distance constraints obtained from CYANA macro caliba or calibration can be used as input files.
- Distance constraints obtained from CYANA macro peak calibrate should have the pseudo-atom correction run before used as input files, run following commands under CYANA:
read upl final.upl
distance correct write upl final_corrected.upl This, however, will not add corrections for multiplicity!
- d2X, to run: d2X cyana2.aco cns.tbl This will convert angle constraints from CYANA format to CNS format. If you don't have any dihedral angle constrains, create an empty file.
Both p2X and r2X require a translation table file atomtransC.tbl.
If have used simplified pseudoatom names (H* instead of Q*) with option pseudo=2 in CYANA 2.1, you may first want to them before running p2X and r2X:
pseudo=0 read upl noe.upl read pdb cyana2.pdb write upl out.upl write pdb out.pdb all
Note that pseudo=0 should be set before loading PDB and constraint files.
Conversion from DYANA
Use CYANA macro MigrateFromDyanaCyana1.cya to convert of data files from standard CYANA1.x or DYANA nomenclature to the standard IUPAC nomenclature used by CYANA, then do then same thing as above.
translate dyana # use Cyana 1.x/Dyana nomenclature read seq demo-dyana.seq # read sequence from Cyana 1.x/Dyana read upl demo-dyana.upl unknown=warn # read upper distance limits from Cyana 1.x/Dyana read aco demo-dyana.aco unknown=warn # read angle restraints from Cyana 1.x/Dyana translate off # return to standard (IUPAC) nomenclature write demo-cyana.seq # save sequence in standard Cyana format write upl demo-cyana.upl # save upper distance limits in standard Cyana format write aco demo-cyana.aco # save angle restraints in standard Cyana format
Running CNS using WaterRefCNS script at CABM
The WaterRefCNS script, developed at Rutgers University by Roberto Tejero, performs many of the above CNS water refinement protocols in an easy to use script with many user controled options.
To see a list of options type the command:
WaterRefCNS
You will see a list of options and how the program is used.
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
WaterRefCN -- a tool to launch structure refinement with water
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Usage: /farm/software/WaterRefinement_cns/WaterRefCNS -na NamProt -que [PBS|No] -pro [xpl|cns]
** Full set of options [not all are needed for a perfect run, use defaults]
-na NamProt Name of the protein NOTE: this is mandatory, no default value
-qu [PBS|NO] Que system to use, one of PBS or NOQUE def NOQUE
-pr [cns|xpl] Protocol to use, X-POR, CNS def cns
-av [center|sum] average for the distance restraints (for CNS, XPLOR) def sum
-ci n1,n2 CIS pep info (PRO res num) in comma separated list, i.e 56,89
-ss n1-n2,n3-n4 S-S bridge info in comma separated, dash-separated pairs, i.e. 2-24,30-40
-he Display this help message
-hisd n1,n2 HISD pep info (HIS res num) in comma separated list, i.e 12,43
-hise n1,n2 HISE pep info (HIS res num) in comma separated list, i.e 25,67
-heat N number of cycles in heating stage, default is 200
-hot N number of cycles in HOT stage, default is 1000
-cool N number of cycles for cooling stage, default is 100
-sc N Scale all terms (noe, dihe, hbond) by xN times def 1
-par string Choice of nonbonded params, one of OPLSX|PARAM19|PARMALLH6|PROLSQ|CONTACT
default is OPLSX
-seed <int> Seed for random number generator, default 31415
Examples:
/farm/software/WaterRefinement_cns/WaterRefCNS -na SR358 -que no -pr cns -ci 22
Given a protein name KKK, here are the input files required to run WaterRefCNS:
- pdb coordinates (in a single file) prepared in Xplor format using PdbStat. The name of the file should be: KKK.pdb
- an noe.tbl file in CNS format. You can convert CYANA a final.upl file using PdbStat. We typically add 10-15% to the upper limit and make the lower limit vdw. The name of this file should be: KKK_noe.tbl
- a dihe.tbl file in CNS format. Again, you can convert a CYANA .aco file using PdbStat. The name of this file should be: KKK_dihe.tbl
- an hbond.tbl file in CNS format (optional). The name of this file should be: KKK_bhond.tbl
In order to run WaterRefCNS on the CABM-NMR cluster:
- Log into hummer or master3
- go to the directory with the above 4 files
- type:
/farm/software/WaterRefinement_cns/WaterRefCNS -na KKK -que PBS
and that's it. If you have some CIS residue you add -ci NUMRES to the options above. If you want to have better vdw violation, you can add -par PARAM19 to the options above.
Future work:
Currently, the WaterRefCNS script does not support refinement with RDC's.
WaterRefCNS at SUNY Buffalo
The CABM water refinement package was customized to run on computers at UB:
- Fixed a bug in WaterRefCNS so that the assembled refined PDB structure file has a .pdb extension.
- PBS queue submission code in WaterRefCNS modified for use on U2 cluster
- Added -hisd and -hise options to use neutral histidine isoprotomers. This required modification of WaterRefCNS, cns_refine_h2o.inp and generate_h2o.inp.
- Fixed a bug in the protein topology file topallhdg5.3.pro. The line atom CD1 type=CR1E charge=0.130 end in the HISE patch section was removed.
- Fixed a bug that caused the order of conformers to be changed in the output PDB file by replacing the line $WATREFLIB/Agrupa *.pdb > All_${Name}_cns.pdb with $WATREFLIB/Agrupa `ls resa_?.pdb ; ls resa_??.pdb` > All_${Name}_cns.pdb in the WaterRefCNS script.
The modified package is installed in /nsm/chem/cen2/HTP2/3_src/WaterRefinement_cns on the local server and in /san/projects1/szypersk/src/WaterRefinement_cns on the U2 Linux cluster. Make sure that this directories is included in your path on spins* workstations and U2, respectively.
For reference, the download location is here: WaterRefinement_UB.tar.Z
The required files are:
- PDB coordinates (with atom names in XPLOR/CNS format)
- NOE constraints (in XPLOR/CNS format)
- Dihedral angle constraints (in XPLOR/CNS format)
- Hydrogen bond constraints (in XPLOR/CNS format)
Assuming that your coordinate file is called KKK.pdb, the constraint files should be named KKK_noe.tbl, KKK_dihe.tbl and KKK_hbond.tbl. If you don't have dihedral angle and/or hydrogen bond constraints, then create the corresponding empty files.
You may need to add a line like nassign=1000 at the top of the constraint files. The value should be equal to or greater than the number of constraints in the file.
WaterRefCNS assumes "sum of r-6" averaging by default. When using "center"-averaged UPLs (e.g. from caliba) add -av center when starting WaterRefCNS. For information on averaging conventions for calibration in CYANA see NOE Calibration in CYANA
It is recommended to run it on U2 Linux cluster. Examples:
WaterRefCNS -na KKK -que PBS
WaterRefCNS -na KKK -que PBS -ci 21,49
residues 21 and 49 are cis-ProWaterRefCNS -na KKK -que PBS -hise 40,82 -hisd 32,65
residues 40 and 82 are ε-protonated neutral His
residues 32 and 65 are δ-protonated neutral HisWaterRefCNS -na KKK -que PBS -av center
for use with center-averaged calibration
To run on spins* Linux workstations without queue system use -que NO option instead of -que PBS option. The default is no queue system, so you can omit the -que option altogether.
Running WaterRefCNS
- Convert the coordinate and constraint files as described above.
- Copy the converted files into a CNS working directory on the U2 cluster or a workstation (e.g. structure/cns/calc1).
- Run the WaterRefCNS with the proper arguments.
- If the calculation was successful, the refined coordinate file will be stored in the refinedPDB subdirectory. Check the BeSureToREADME for details.
Running CNS "manually"
Go to ~1_projects/targetID/structure/cns/ directory and cns_scripts sub directory, follow the procedures as describe below.
1. Generating MTF topology file from a PDB file.
modify the header of the input file generate_mtf.inp and then run cns < generate_mtf.inp > mtf.log.
- Modify input files path and name base on individual target as follows:
...
{in} pdb_file="/san/user/gliu2/u2/cns/cns/convertion_scripts/ufc1_1.pdb";
{in} param_file="/san/user/gliu2/u2/cns/cns/cns_scripts/parallhdg5.3C.pro";
{in} topol_file="/san/user/gliu2/u2/cns/cns/cns_scripts/topallhdg5.3.pro";
{in} plink_file="/san/user/gliu2/u2/cns/cns/cns_scripts/protein.link";
{out} struct_file="ufc1_1.mtf";
...
- Requires these files:
- parallhdg5.3C.pro: CNS parameter file
- topallhdg5.3.pro: CNS topology file
- protein.link: CNS peptide linkage definition file
- For cis-Proline, define the residue Id of the residue prior to cis-Proline by additional lines in generate_mtf.inp before the WRITE command
patch cisp
reference=nil=( resid 90 )
end
- If you get an error message on the atom type, you also can try
patch cipp
reference=nil=( resid 90 )
end
- For neutral Nε- and Nδ-protonated histidines add these lines in generate_mtf.inp before the WRITE command
patch hise
reference=nil=( resid 40 )
end
patch hisd
reference=nil=( resid 65 )
end
- For dimer refinement, add a line containing only TER between two monomer unit coordinates in the PDB conformer that is defined in generate_mtf_cis.inp as input file.
2. Rebuilding hydrogen atom positions for each structure.
Modify input file rebuild.inp and then run cns < rebuild.inp > rebuild.log
- Modify input files path and name base on induvidual target as follows:
... evaluate ($mtf_file="ufc1_1.mtf") evaluate ($pdbname_in="../convertion_scripts/ufc1") evaluate ($pdbname_out="ufc1_rb") evaluate ($number_of_struct= 20 ) ... evaluate ($topol_p_file="/san/user/gliu2/u2/cns/cns/cns_scripts/topallhdg5.3.pro") evaluate ($param_p_file="/san/user/gliu2/u2/cns/cns/cns_scripts/parallhdg5.3C.pro") ...
- Requires these files:
- parallhdg5.3C.pro: CNS parameter file
- topallhdg5.3.pro: CNS topology file
3. Refining structures with explicit water.
- Running CNS on workstation with single processor, modify input file re_h2oc.inp and then run cns < re_h2oc.inp > h2o.log
- Modify input files path and name base on induvidual target as follows:
... evaluate ($mtf_file="ufc1_1.mtf") evaluate ($noe_file="../noe.tbl") evaluate ($dihe_file="../aco.tbl") evaluate ($hb_file="../hbond.tbl") evaluate ($pdbname_in="ufc1_rb") evaluate ($pdbname_out="ufc1_ref") evaluate ($number_of_struct= 20 ) ...
- Modify the weight of energy contribution part if necessary, e.g. use 2x to 10x of the following scales:
scale ambi 50 scale dist 50 scale hbond 50 end restraints dihedral scale=200 end
- Requires
- parallhdg5.3C.pro: CNS parameter file
- topallhdg5.3.pro: CNS topology file
- topallhdg5.3.sol: CNS solvent topology file
- parallhdg5.3.sol: CNS solvent parameter file
- boxtyp20.pdb: Water box
- generate_water.cns: CNS protein solvation script
- print_coorheader.cns: CNS script
- Run CNS on workstation with multiple processors.
CNS will refine each conformer independently and in parallel, it will be much faster if 20 conformer were refined in parallal at u2.ccr.buffalo.edu.
1. Run cns_convertion as descibed above
2. Go to cns_scripts directory, modify input file generate_mtf_cis.inp and then run =cns < generate_mtf_cis.inp = as described above
3. Run macro getfil
cp ../convertion_scripts/noe.tbl . cp ../convertion_scripts/hbonds.tbl . cp ../convertion_scripts/aco.tbl . cp ufc1_1.mtf com/.
4. Modify input file rebuild.inp and then run cns < rebuild.inp as described above.
5. Modify input file re_h2oc.inp as described above, remember to set $number_of_struct = 20.
6. Run macro subaba
#bash for i in 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 do mkdir ref$i cd ref$i cp ../com/* . cp ../jan24/ufc1_rb_$i.pdb ufc1_rb_1.pdb qsub cns.sc cd .. donedone
where cns.sc is a PBS macro used to submit CNS job by using PBS:
#!/bin/csh #PBS -m e #PBS -q short_c #PBS -l nodes=1:GM:ppn=1 #PBS -l walltime=00:56:00 #PBS -o out1 #PBS -j oe #PBS -N clean1 cd $PBS_O_WORKDIR echo "working directory = "$PBS_O_WORKDIR set NN = `cat $PBS_NODEFILE | wc -l` echo "NN = "$NN # Run Job cns < re_h2oc.inp echo "ALL Done!"
7. Run getpdb to collect refined structures.
#bash for i in 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 do cd ref$i cp ufc1_ref_1.pdb ../pdb/ufc1_ref_$i.pdb cp ufc1_ref_1.vio ../pdb/ufc1_ref_$i.vio cd .. done
8. Run script Agrupa to put all 20 conformers into a single PDB file.
Agrupa ufc1_ref*.pdb > ufc1all.pdb
Running CNS for Proteins with Metal Ions
The CNS refinement for proteins with metal ions can also be performed with the new WaterrefCNS script, using the appropriate metal ion nomenclature in both the pdb coordinates and distance constraints.
For manual CNS refinement please do the refinement as following:
1. Set environment for CNS1.1 by run source /farm/users/gliu/alias.cns
alias cns1 /farm/software/cns/cns_solve_1.1/intel-i686-linux_g77/bin/cns
setenv CNS_TOPPAR /farm/data/gliu/cns1/
In addition to the topology and parameter files, the metal ion parameter file ion.top is required. An example can be found in "/farm/users/gliu/projects/cns_cuttha_cis" with all required input files. 2. Prepared required files as described above (final.tbl, final cns format PDB files and put in xplorPDB dir with name as sa_#.pdb) except the PDB file should include the metal ions with format according to CNS library ion.top. cp sa_1.pdb as template.pdb, input files for creating mtf file Note that alignment is important. eg:
ATOM 1249 OT2 ALA 83 69.296 13.232 5.744 1.00 0.00 ATOM 1250 ZN+2 ZN2 150 63.086 13.789 -10.407 1.00 0.00 zinc
3. Run generate_h2o.inp once to create temp_h2o.pdb and temp_h2o.mtf. The extra proton atom in the ligand residues, eg. HIS HD1 or CYS S, are removed by editing the generate_h2o.inp; cis proline is also defined here in the generate_h2o.inp (resid is the residue number prior the proline).
{* any special prosthetic group patches can be applied here *}
{===>}
delete select (name hg and resname cys and resid 61) end
delete select (name hg and resname cys and resid 85) end
delete select (name hd1 and resname his and resid 46) end
delete select (name he2 and resname his and resid 83) end
patch cisp
reference=1=( resid 13 )
end
4. Edit generate_1.inp to remove the extra proton as did above.
5. Run generate_20.com, this will run generate_#.inp 20 times, updating each pdb number and this creates cnsPDB/sa_cns_#.pdb
6. Edit and run re_h2o_cu.inp, the refined pdb is kept in refinedPDB, or
7. Use subcns to submit cns refinement by using PBS: eg, type "sh subcns". Before run subcns , make a folder " com" contains the following file. Type getpdb to get refined pdb files in refinedPDB after it finished.
- cns.sc: PBS submission
- cutc_h2o.mtf: mtf file created as descrive above
- topology and parameter files: parallhdg5.3C.pro, parallhdg5.3.pro, topallhdg5.3.pro
- re_h2o_cu.inp: input file for cns refinement
Running CNS with RDC Constraints
Under construction
Running CNS refinement on Homodimers
Under construction
CNS Refinement with RDC constraints, metal ions using FMCGUI at UofT
<meta content="text/html; charset=utf-8" http-equiv="CONTENT-TYPE"><title></title> <meta content="OpenOffice.org 3.2 (Linux)" name="GENERATOR"><style type="text/css"> </style>
Step1. Input files
In order to perform water refinement using FMCGUI the following files should be prepared/available:
- structure file
- distance constraints file
- dihedral angles constraints file
User can include in the refinement the following additional constrains/data as well:
- hydrogen bond constrains file
- RDC data collected in different medias; (up to 3 medias are allowed)
- Zinc ions
- Structure file and constraints file format.
It is expected that the structural, distance constraints, and dihedral angle constraints files are available as output of CYANA2.1 /CYANA3.0 structure calculations. The format of these files correspond to the format of'final.pdb', 'final.upl', and 'final.aco'ouput files. At the same time, these files could be loaded in FMCGUI in CNS/XPLOR format as well. It should be noted that structural file should contain key word MODEL that separate lines corresponding to different structural models.
2. RDC data format
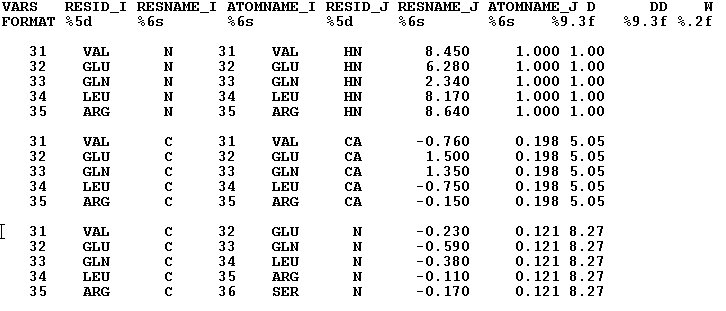
The RDC data can be used in CYANA or PALES formats. One can use RDC data collected in up to 3 media. All data from the same media should be included in the same file. When data are prepared in PALES format than a file should contain data only from one media. When using CAYANA format, data from different media can be included in one file.(See Examples r1,and r2)
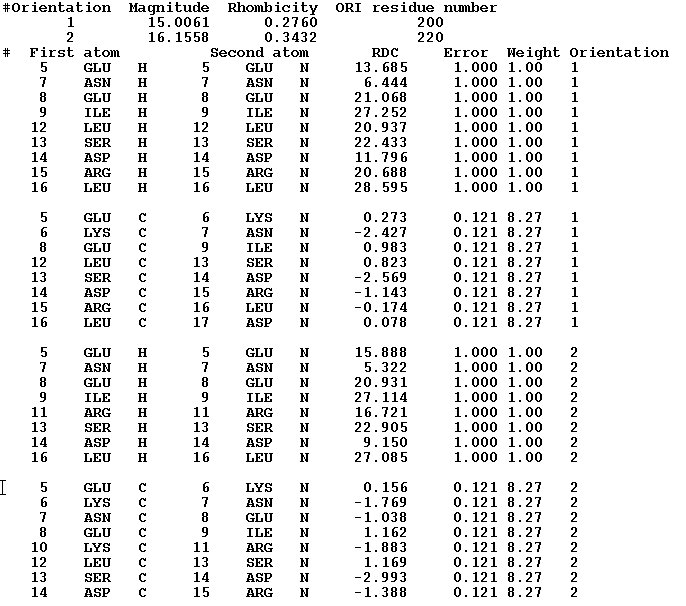
Example r1.RDC data in CYANA3 format.
This example present file with HN and NC rdc's measured in 2 different media.
Example r2. RDC data in PALES format.
This example present file with HN, CCA, and NC rdc's measured in 1 media.
3. Zinc ligands file format.
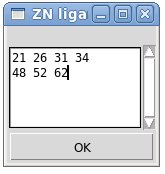
The Zinc-ligands file contains list of residues that coordinate each of Zinc ions present in the structure. There could be 3 or 4 Zinc ligands for one Zinc ion. Possible types of residues that coordinate Zinc ion are CYS, HIS, ASP, and GLU. (See Example z1)
Example z1. Zinc- ligands file.
This file indicate that there are 2 Zinc ions, one Zinc is coordinated by 4 residues 21, 26, 31, and 34 while the other Zinc is coordinated by 3 residues 48, 52, and 62
This Zinc-file could be created using FMCGUI command [Structure>Add ZN ligands]:
User have to type IDs of residues that ligate zinc ion into the popped up entry window “ZN ligands” (see Figure). If there are a few zinc ions, information for each ion should be provided in a separate row.
Step 2. Setting up water refinement calculations using FMCGUI.
2.1 Open new project [Project>new] or load the old one [Project>load]
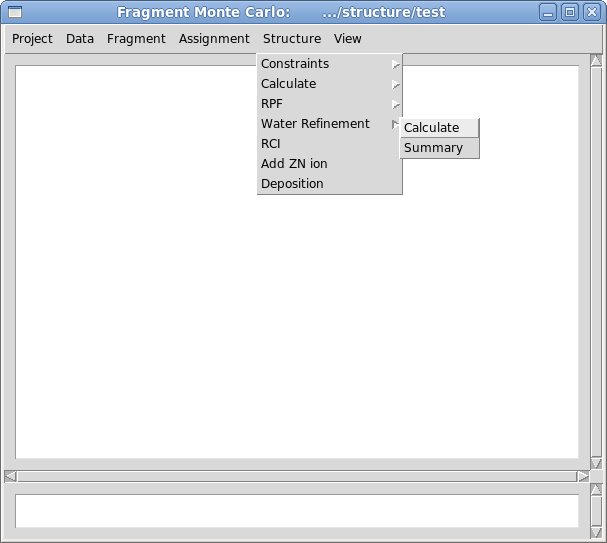
2.2 Select command [Structure>Water Ferinement>calculate]
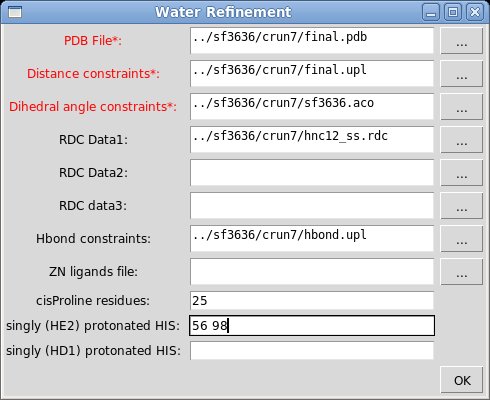
The popped up “Water Refinement” window were user is asked to select a number of files with coordinates and constrains (see Figure). The entry fields marked in read (PDB file, Distance constraints, and Dihedral angle constraints) should be specified while the others could be left blank. In addition to coordinates and constraints, user can indicate cisProline residues (if there are any) and to specify singly protonated HIS residues that are double protonated by default.
By pressing OK button in “water Refinement” window a new “Parameters” window
will pope up. User have to specify
- name of the water refinement directory
- flag for distance constraints transformation (could be 1 or 2)
- force
constants for RDC constraints (if there are any) on the final stage of the refinement protocol
Pressing OK button starts the setup of water refinement calculations using provided information the specified directory.
The set up consists of the following operations:
- Splitting structure file in a number of separate files for each model. The atom names are changed from CYANA to CNS nomenclature.
- Transforming dihedral angle constraints from CYANA to CNS format. (If constraints are provided in CNS format then no transformation is performed).
- Transforming distance NOE constraints from CYANA format to CNS format. This transformation includes changing atom names from CYANA to CNS nomenclature. If the flag for distance constraints transformation is set to 2 than no change in the upper distance limit is performed. If the flag is set to 1 (default value), then the upper distance limits are modified by classifying each constraint in one of the 4 classes – strong, medium, weak and very weak.
- Transforming Hbond constraints (if any) from CYANA format to CNS format
- Generating mtf-file for CNS calculations
- “Repairing”
each input structure in accord with mtf-file
- Generating RDC constraints, a separate file for each media
- Estimating Da and R of alignment tensor for each media. This is performed by fitting RDC data to each model structure using program PALES, with subsequent averaging of the fitting results.
- Generating
CNS script “re_h2o*_tmpl.inp” for the refinement calculations
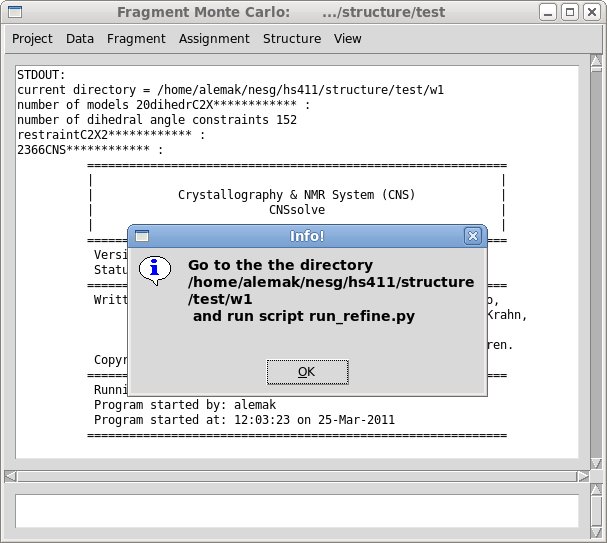
Step 3. Water refinement calculations using CNS.
The calculations could be perform by running python script “run_refine.py”.
This script will start N jobs in parallel, were N is the number of
structural models to refine. The number of structures and the number of processor to use in the calculations can be specified by making modification of the top lines in script “run_refine.py”. If user want to reset the values for some refinement parameters, for example the values of Da and R, it could be done by modifying corresponding lines in the header of the CNS script “re_h2o*_tmpl.inp”.
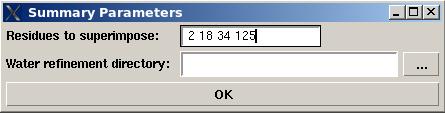
Step4. Summaraizing results of refinement using.
Open new project [Project>new] or load the old one [Project>load]
Select command [Structure>Water Refinement>summary]
In the popped up “Water refinement summary” window user will be asked to specify residues used for structure superposition and to select the water refinement directory “WATDIR” were the refinement calculations were made.
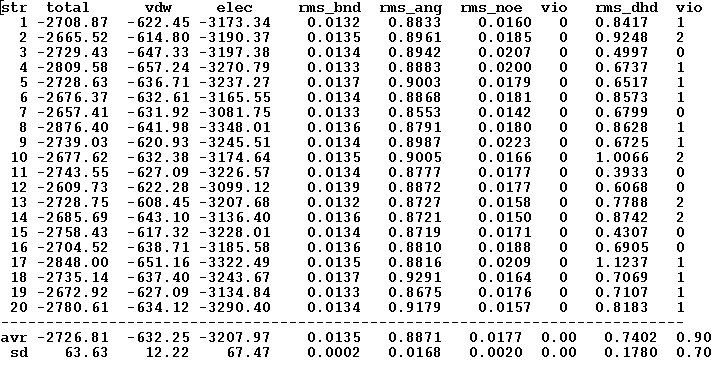
The refined structural models will be superimpose and combined in one file. The created summary reports values of different energy components and constraint violation statistic for each structural model. A new directory “WATDIR_results” will be created. The directory contains final superimposed coordinates, distance and dihedral angle constrains in a format suitable for PDB deposition, summary and constraint violations reports, files“final_cns_summary” and “final_violations” respectively. (See examples S1 and S2)
Example S1</u</font</font><span lang="en-US" />.<font size="2" style="font-size: 11pt;"</font<span style="font-weight: normal;" /></font></font>
File “final_cns_summary”
The file shows the total energy, energy components, and number of constraints violations for each refined structure as well as corresponding average over all structures values.
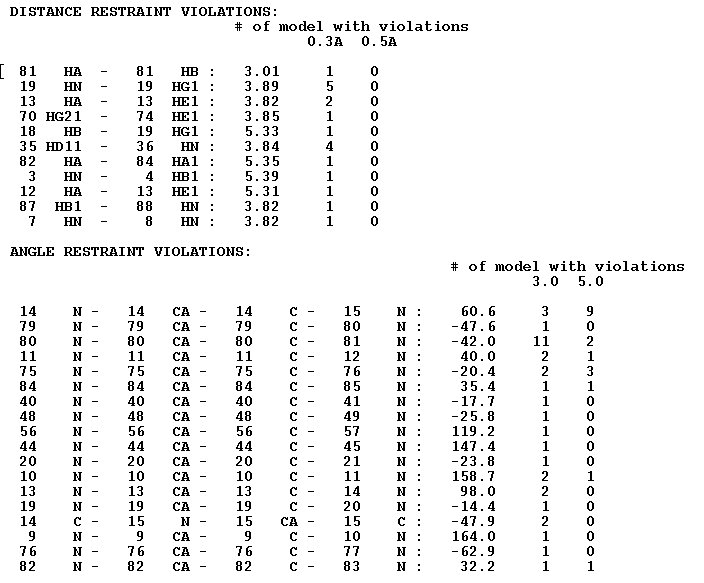
<u>Example s2.</u>File “final_violations”
The file contains a list of all constraints violated in at least one structural model. For each such a constraint the the number of f structural model with tviolations of the constraint is shown.
</meta></meta>
Files for Download
- atomtransC.tbl: atom name translation table
- Agrupa: script for concatenating pdb coordinate files into one file
- parallhdg5.3C.pro: CNS parameter file
- topallhdg5.3.pro: CNS topology file
- topallhdg5.3.sol: CNS solvent topology file
- parallhdg5.3.sol: CNS solvent parameter file
- boxtyp20.pdb: Water box
- protein.link: CNS peptide linkage definition file
- generate_water.cns: CNS protein solvation script
- print_coorheader.cns: CNS script
- cns_cabm.tar.gz: CABM CNS refinement protocol
- WaterRefinement_UB.tar.Z: CNS water refinement package - SUNY Buffalo
- cyana2cns.cya: CYANA script to convert coordinates and constrains to XPLOR/CNS format
- Cu_cPro_cns_example.tar.gz: Example of CNS refinment for protein with Cu and cis-Pro
- zn.tar: Exampleof CNS refinment for protein with zn (from Alex Lemark)
- er317_2zn_CNSw_example.tar.gz: Example of CNS refinment for protein with zn (from Theresa Ramelot)
References