AutoStructure: Difference between revisions
Jump to navigation
Jump to search
Control File
(Created page with '== '''Introduction''' == == '''Getting Started''' == === Input Files === === Graphical User Interface === === Control File === == '''Files for Download''' == == '''Re…') |
No edit summary |
||
| Line 4: | Line 4: | ||
=== Input Files === | === Input Files === | ||
=== Graphical User Interface === | === Graphical User Interface === | ||
=== Control File<br> === | |||
=== Staring a Calculation === | |||
The user can start a calculation from either the graphical user face or the command line. | |||
1. Launching a calculation from the GUI. | |||
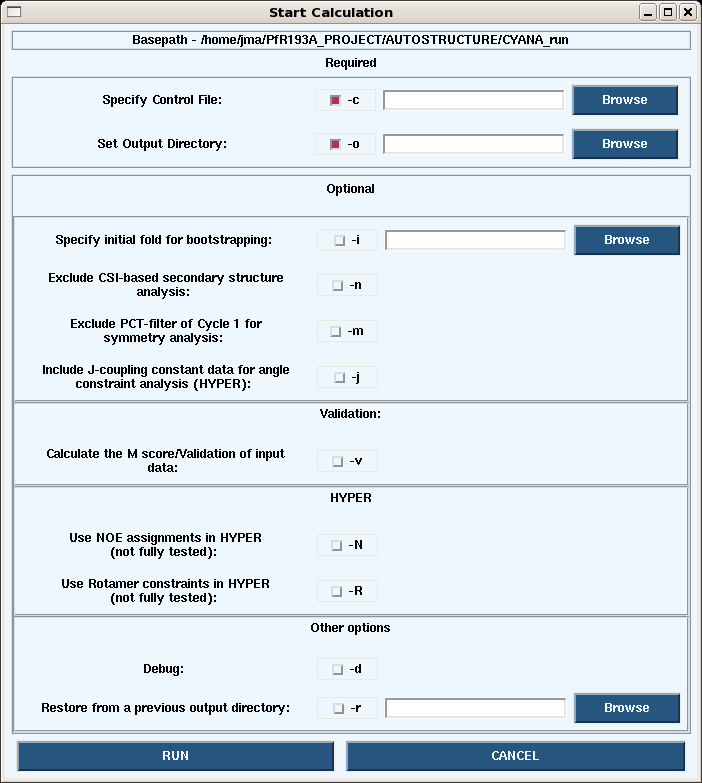
*Under the Autostructure pull-down on the main page, choosing Start -> Calc opens the dialogue box below. | |||
[[Image:AS_startCalc.png]] | |||
*Launching calculations from the GUI uses is generally slow since only the processors on your own machine are used. To speed up the calculations use the command line on a cluster. | |||
2. Launching a calculation from the command line. | |||
*Login in the a cluster. At CABM we use AutoStructure version 2.2.1 on hummer. | |||
*A simple command line run can be started as follows: | |||
<pre>/farm/software/AutoStructure/AutoStructure-2.2.1/bin/autostructure -c controlfile_CYANArun -o testCYANArun.out -v</pre> | |||
*Running the autostructure command gives the following options (like those avaliable in the GUI): | |||
<pre> AutoStructure/RPF Version 2.2.1 Copyright(C) 2007 | |||
Center for Advanced Biotechnology and Medicine (CABM) | |||
Rutgers University | |||
Options: | |||
-c control_file Required | |||
-o output_dir Required | |||
-d For debug | |||
-h Help | |||
-m Exclude PCT-filter of Cycle1 for symmetry analysis | |||
-n Exclude CSI-based secondary structure analysis | |||
-i structure_file inital fold for bootstrapping | |||
-j Include J-coupling constant data for angle constraint analysis (HYPER) | |||
-k float_number Calibration coefficient | |||
-N Include NOE assignments in HYPER caluclation (under development) | |||
-q structure_file AutoQF-Calculate the F and DP scores of the input structure_file (IUPAC naming) | |||
-r path Restore from a prior outout_dir | |||
-R Include rotamer constraints in HYPER calculation (under development) | |||
-v Calculate the M score and average shifts | |||
</pre> | |||
== '''Files for Download''' == | == '''Files for Download''' == | ||
== '''References''' == | |||
== '''References''' == | |||
-- JimAramini - 07 Nov 2009 | |||
Revision as of 19:38, 7 November 2009
Introduction
Getting Started
Input Files
Graphical User Interface
Control File
Staring a Calculation
The user can start a calculation from either the graphical user face or the command line.
1. Launching a calculation from the GUI.
- Under the Autostructure pull-down on the main page, choosing Start -> Calc opens the dialogue box below.
- Launching calculations from the GUI uses is generally slow since only the processors on your own machine are used. To speed up the calculations use the command line on a cluster.
2. Launching a calculation from the command line.
- Login in the a cluster. At CABM we use AutoStructure version 2.2.1 on hummer.
- A simple command line run can be started as follows:
/farm/software/AutoStructure/AutoStructure-2.2.1/bin/autostructure -c controlfile_CYANArun -o testCYANArun.out -v
- Running the autostructure command gives the following options (like those avaliable in the GUI):
AutoStructure/RPF Version 2.2.1 Copyright(C) 2007
Center for Advanced Biotechnology and Medicine (CABM)
Rutgers University
Options:
-c control_file Required
-o output_dir Required
-d For debug
-h Help
-m Exclude PCT-filter of Cycle1 for symmetry analysis
-n Exclude CSI-based secondary structure analysis
-i structure_file inital fold for bootstrapping
-j Include J-coupling constant data for angle constraint analysis (HYPER)
-k float_number Calibration coefficient
-N Include NOE assignments in HYPER caluclation (under development)
-q structure_file AutoQF-Calculate the F and DP scores of the input structure_file (IUPAC naming)
-r path Restore from a prior outout_dir
-R Include rotamer constraints in HYPER calculation (under development)
-v Calculate the M score and average shifts
Files for Download
References
-- JimAramini - 07 Nov 2009