CARA One Letter File To Seq File: Difference between revisions
(Created page with 'Generating XEASY sequence file Before you create a project you may need to generate an XEASY sequence file. Run "OneLetterFileToSeqFile" Lua script from Terminal -> !OneLette…') |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
Generating XEASY sequence file | Generating XEASY sequence file | ||
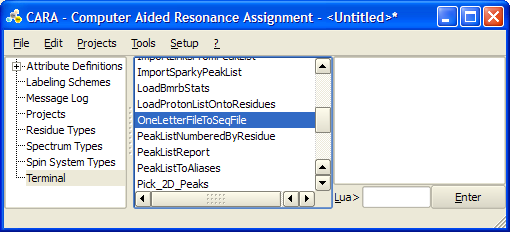
Before you create a project you may need to generate an XEASY sequence file. Run "OneLetterFileToSeqFile" Lua script from Terminal -> | Before you create a project you may need to generate an XEASY sequence file. Run "OneLetterFileToSeqFile" Lua script from '''Terminal -> OneLetterFileToSeqFile'''. | ||
Creating XEASY sequence file: | Creating XEASY sequence file: | ||
[[Image:CARA One Letter to Seqfile.png]] | |||
In the unlikely case that your template does not contain this script, you can get it here: [http://www.cara.ethz.ch/Wiki/CALUA http://www.cara.ethz.ch/Wiki/CALUA] | |||
This script will open a browser window, where you can select the file containing protein sequence as a single line of one-letter codes. This line you can copy directly from the target entry in SPINE. The filename must be in the form of foo.aa. | |||
Then the script will ask you for the name of the output foo.seq file | |||
-- Main.AlexEletski - 07 Mar 2007 | It should be possible to use the sequence produced by UBNMR, but you will need to remove all SRD spin systems from it. | ||
-- Main.AlexEletski - 07 Mar 2007 | |||
[[Category:CARA]][[Category:Resonance_Assignment]] | |||
Latest revision as of 22:40, 9 November 2009
Generating XEASY sequence file
Before you create a project you may need to generate an XEASY sequence file. Run "OneLetterFileToSeqFile" Lua script from Terminal -> OneLetterFileToSeqFile.
Creating XEASY sequence file:
In the unlikely case that your template does not contain this script, you can get it here: http://www.cara.ethz.ch/Wiki/CALUA
This script will open a browser window, where you can select the file containing protein sequence as a single line of one-letter codes. This line you can copy directly from the target entry in SPINE. The filename must be in the form of foo.aa.
Then the script will ask you for the name of the output foo.seq file
It should be possible to use the sequence produced by UBNMR, but you will need to remove all SRD spin systems from it.
-- Main.AlexEletski - 07 Mar 2007