Cofactor optimization: Difference between revisions
mNo edit summary |
mNo edit summary |
||
| Line 12: | Line 12: | ||
=== Addition and removal of Zinc from ''Haemophilus influenzae'' IscU === | === Addition and removal of Zinc from ''Haemophilus influenzae'' IscU === | ||
As many as 30% of proteins are predicted to be metal binding proteins and many of these may require metal for proper folding. In fact, a recent analysis of the PDB revealed that 14% of non-redundant entries contain stoichiometric amounts of six transition metals: Mn, Fe, Co, Ni, Cu or Zn <ref>Shi, W., Zhan, C., Ignatov, A., Manjasetty, B.A., Marinkovic, N., Sullivan, M., Huang, R., and Chance, M.R. (2005). Metalloproteomics: high-throughput structural and functional annotation of proteins in structural genomics. Structure 13, 1473-1486.</ref> | As many as 30% of proteins are predicted to be metal binding proteins and many of these may require metal for proper folding. In fact, a recent analysis of the PDB revealed that 14% of non-redundant entries contain stoichiometric amounts of six transition metals: Mn, Fe, Co, Ni, Cu or Zn <ref>Shi, W., Zhan, C., Ignatov, A., Manjasetty, B.A., Marinkovic, N., Sullivan, M., Huang, R., and Chance, M.R. (2005). Metalloproteomics: high-throughput structural and functional annotation of proteins in structural genomics. Structure 13, 1473-1486.</ref> Of particular interest is zinc, which is the second most abundant transition metal in living organisms after Fe <ref>Lipscomb, W.N., and Strater, N. (1996). Recent Advances in Zinc Enzymology. Chem Rev 96, 2375-2434.</ref>. The extent is highlighted in a recent bioinformatics analysis of the human proteome in which it was determined that as much 10% of the proteome is zinc-bound <ref>Andreini, C., Banci, L., Bertini, I., and Rosato, A. (2006). Counting the zinc-proteins encoded in the human genome. J Proteome Res 5, 196-201.</ref>. In biology zinc is commonly assuming a structural role (structural zinc), and less commonly, a catalytic role <ref>Passerini, A., Andreini, C., Menchetti, S., Rosato, A., and Frasconi, P. (2007). Predicting zinc binding at the proteome level. BMC Bioinformatics 8, 39.</ref>. Structural zinc is tetrahedrally coordinated by the side-chains of a small subset of amino acids. The coordinating atoms are in-turn are stabilized by a hydrogen bonding network with amino acids in an outer coordinating sphere. | ||
<br> In 2006 Passerini et al., developed a de novo predictor of zinc-binding called ZincFinder <ref>Passerini, A., Punta, M., Ceroni, A., Rost, B., and Frasconi, P. (2006). Identifying cysteines and histidines in transition-metal-binding sites using support vector machines and neural networks. Proteins 65, 305-316</ref>. The algorithm uses sequence specific information and is trained on 230 zinc binding chains to recognize position specific evolutionary profiles. The algorithm also considers global parameters, such as chain length and amino acid composition, as well as local parameters, such as the distance between potential ligands. ZincFinder has a reported precision of 73% and a recall of 61%. | |||
<br> In 2006 Passerini et al., developed a de novo predictor of zinc-binding called ZincFinder <ref>Passerini, A., Punta, M., Ceroni, A., Rost, B., and Frasconi, P. (2006). Identifying cysteines and histidines in transition-metal-binding sites using support vector machines and neural networks. Proteins 65, 305-316 | |||
<br> | <br> | ||
ZincFinder was used to analyze 36,761 full length NESG targets with sequence lengths of less than or equal to 1000 amino (http://zincfinder.dsi.unifi.it/). A protein was strongly suspected to bind zinc if three or more Cys, His, Asp or Glu residues were predicted to bind zinc with a probability equal to or greater than 50%; 3,137 targets (8.5% of studied targets) were found that met this criteria. ZincFinder predicts that 24 of the 691 targets structured by the NESG possesses a zinc binding site. Eighteen of the twenty four targets have been deposited with coordinates for their bound zinc atoms. The remaining 6 targets may actually be zinc binding proteins where the zinc was not introduced during fermentation or lost during purification. | ZincFinder was used to analyze 36,761 full length NESG targets with sequence lengths of less than or equal to 1000 amino ([http://zincfinder.dsi.unifi.it/]). A protein was strongly suspected to bind zinc if three or more Cys, His, Asp or Glu residues were predicted to bind zinc with a probability equal to or greater than 50%; 3,137 targets (8.5% of studied targets) were found that met this criteria. ZincFinder predicts that 24 of the 691 targets structured by the NESG possesses a zinc binding site. Eighteen of the twenty four targets have been deposited with coordinates for their bound zinc atoms. The remaining 6 targets may actually be zinc binding proteins where the zinc was not introduced during fermentation or lost during purification. | ||
<br> | <br> | ||
Revision as of 15:17, 17 December 2009
Often an NMR sample can be improved by adding an appropriate cofactor such as a metal or a small organic molecule.
Here we show two examples:
- Addition of Zn to IscU
- Addition of CoA to YhhK
Addition and removal of Zinc from Haemophilus influenzae IscU
As many as 30% of proteins are predicted to be metal binding proteins and many of these may require metal for proper folding. In fact, a recent analysis of the PDB revealed that 14% of non-redundant entries contain stoichiometric amounts of six transition metals: Mn, Fe, Co, Ni, Cu or Zn [1] Of particular interest is zinc, which is the second most abundant transition metal in living organisms after Fe [2]. The extent is highlighted in a recent bioinformatics analysis of the human proteome in which it was determined that as much 10% of the proteome is zinc-bound [3]. In biology zinc is commonly assuming a structural role (structural zinc), and less commonly, a catalytic role [4]. Structural zinc is tetrahedrally coordinated by the side-chains of a small subset of amino acids. The coordinating atoms are in-turn are stabilized by a hydrogen bonding network with amino acids in an outer coordinating sphere.
In 2006 Passerini et al., developed a de novo predictor of zinc-binding called ZincFinder [5]. The algorithm uses sequence specific information and is trained on 230 zinc binding chains to recognize position specific evolutionary profiles. The algorithm also considers global parameters, such as chain length and amino acid composition, as well as local parameters, such as the distance between potential ligands. ZincFinder has a reported precision of 73% and a recall of 61%.
ZincFinder was used to analyze 36,761 full length NESG targets with sequence lengths of less than or equal to 1000 amino ([1]). A protein was strongly suspected to bind zinc if three or more Cys, His, Asp or Glu residues were predicted to bind zinc with a probability equal to or greater than 50%; 3,137 targets (8.5% of studied targets) were found that met this criteria. ZincFinder predicts that 24 of the 691 targets structured by the NESG possesses a zinc binding site. Eighteen of the twenty four targets have been deposited with coordinates for their bound zinc atoms. The remaining 6 targets may actually be zinc binding proteins where the zinc was not introduced during fermentation or lost during purification.
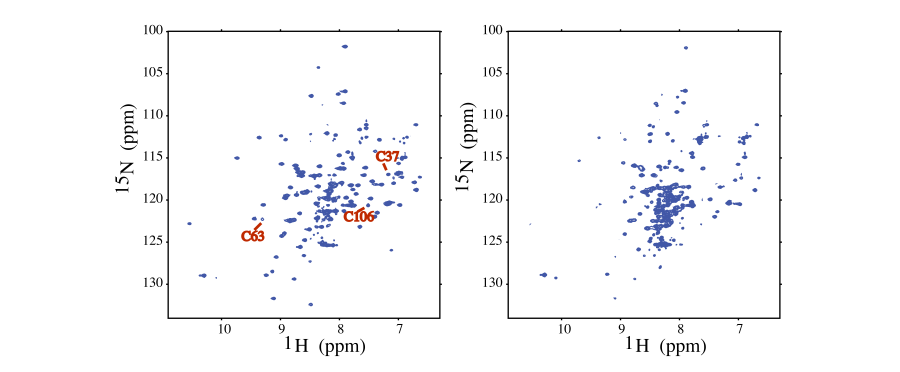
In this example (NESG ID IscU) the effect of metal chelation on protein stability was probed by a 2D 1H–15N HSQC NMR experiment. The well dispersed spectra is indicative of global stability (left), collapsed spectra of unstructured protein (right) [6]. Interestingly, IscU is an Fe-binding protein in vivo, but the metal is replaced by Zn during NMR sample preparation.
Due to the adverse effect of metal chelation on protein structure, and the high prevalence of zinc, it is likely that a subset of NESG targeted proteins with poorly resolved 2D 1H–15N HSQC spectra, may have inadvertently lost structural zinc during the protein sample preparation.
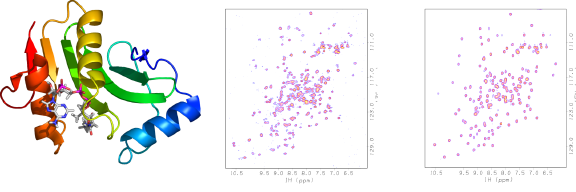
Addition of acetyl CoA to E. coli YhhK
E.coli YhhK (NESG ID ET106) is a putative GCN5-related N-acetyltransferase (NAT) whose substrate is unknown (below, left). The protein purified with partial occupancy of the acetyl CoA binding site, thereby yielding a N-HSQC spectrum during screening with too many peaks (below, middle) because both apo- and holo-protein were present. After additional CoA was added the spectrum showed only one set of peaks (below, right), from ligand-bound protein. The observation of multiple peaks initially also indicates ligand binding in slow exchange on the NMR time scale. Additional CoA was added and the spectrum showed only one set of peaks, from ligand-bound proteins.
References
- ↑ Shi, W., Zhan, C., Ignatov, A., Manjasetty, B.A., Marinkovic, N., Sullivan, M., Huang, R., and Chance, M.R. (2005). Metalloproteomics: high-throughput structural and functional annotation of proteins in structural genomics. Structure 13, 1473-1486.
- ↑ Lipscomb, W.N., and Strater, N. (1996). Recent Advances in Zinc Enzymology. Chem Rev 96, 2375-2434.
- ↑ Andreini, C., Banci, L., Bertini, I., and Rosato, A. (2006). Counting the zinc-proteins encoded in the human genome. J Proteome Res 5, 196-201.
- ↑ Passerini, A., Andreini, C., Menchetti, S., Rosato, A., and Frasconi, P. (2007). Predicting zinc binding at the proteome level. BMC Bioinformatics 8, 39.
- ↑ Passerini, A., Punta, M., Ceroni, A., Rost, B., and Frasconi, P. (2006). Identifying cysteines and histidines in transition-metal-binding sites using support vector machines and neural networks. Proteins 65, 305-316
- ↑ Ramelot, T.A., et. al. (2004) Solution NMR structure of the iron-sulfur cluster assembly protetin U (IscU) with zinc bound the active site. Journal of Molecular Biology, 344, 567-583