GLOMSA
SSA with GLOMSA (GLObal Method for Stereospecific Assignment) Module
Overview
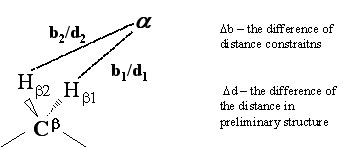
GLOMSA is performed with the preliminary conformers and using all the local and global experimental data. If there are two NOEs of significantly different strength from a given proton to both diastereotopic substituents of a prochiral center and if the distances from the given proton to the two diastereotopic substituents differ consistently in the structures, the stronger NOE can be identified with the diastereotopic substituent that is closer to the given proton.
If |Δb| > cutoff, and |Δd| > threshold, the diastereotopic pair is assigned as:
- input (if Δb and Δd are both either positive or negative)
- reversed (if Δb and Δd have opposite signs)
- GLOMSA usage:
Parameters: atom selection (default: all atoms) cutoff=c (default: 0.4) threshold=t (default: 0.4) fraction=f (default: 100)
Only those atoms are considered, for which the difference between two constraints is larger than cutoff (in Å) and the corresponding average distance difference in the bundle of structure is larger than threshold. The minimal percentage of structures in which the sign of the distance difference must be consistent must be larger than fraction (in percent).
Running GLOMSA
- Copy the latest manual refinement directory.
- Download the sample run_glomsa.cya script
- Start CYANA 2.1 on a workstation and execute run_glomsa.cya. This will generate glomsa.out file.
- Analyze the output (here glomsa.out). For each stereospecific assignments verify that it is justified. Add confirmed assignments to the stereofound.cya file.
- Run a manual structure calculation with the same CALC.cya as before. It will now use updated stereospecific assignments. Delete the glomsa.out file if you don't want it to be appended.
- Repeat steps 3-5 until no new stereospecific assignments can be made.
Important notes:
- GLOMSA requires that you load a structure and UPL constraints. You should also declare the previously established stereospecific assignments (e.g., for VAL and LEU methyl groups) so that these could be used to determine new tereospecific assignments.
- Important! The UPL constraints supplied to GLOMSA must generated before distance modification with distance modify. When you follow the procedure in the manual structure calculation example this would be the $name-in.upl file.
- At this point we usually assign only HB2/3 spins and HA2/3 spins of Gly, HD21/22 of Asn and HE21/22 of Gln. Methyl QG1/2 of Val and QD1/2 of Leu can be assigned at this stage if that has not been done previously with the help of a fractionally 13C-labeled sample. Stereospecific assignments of other spin pair are usually not reliable.
- In the first iteration accept only stereospecific assignments supported by multiple third atoms.
Acceptance criteria for stereospecific assignments from GLOMSA
- No conflicting stereospecific assignments in the output of GLOMSA (for example, both as input and reversed assignments for the same atom pair). Exceptions:
- Some conflicting results may be caused by NOE peaks to QD and QE pseudoatoms of aromatic rings, due to constituent HD1/2 and HE1/2 atoms being so far away from the pseudoatom and slow ring flipping.
- Some conflicting results may be caused by incorrect peak intensities (see next point).
- Verify that the NOE peak intensities (a-b1 and a-b2) are correct, and not biased by overlap with other peaks.
- Verify that the NOE peak intensities have not been misinterpreted (Example: HN-HB2 peak only resolved in n.peaks, and HN-HB3 - only in ali.peaks. Different NOE calibration constants for both peaklists may lead to incorrect stereospecific assignments.)
- Big violations should be detected if the opposite stereospecific assignment is used.
run_glomsa.cya
Here is an example script run_glomsa.cya:
protocol:=glomsa.out read pdb $name.pdb read upl $name-in.upl stereofound # ssa from previous step atom glomsa protocol:=
GLOMSA output
The output of glomsa will look like this:
Atoms residue constraints as input reversed distances assign
sum min sum min num mean assign
HB2/3 ARG 4
H ARG 4 5.50 3.64 0.64 0.00 0.13 0.00 -15 -0.49 ?
HB2/3 LEU 6
HA LEU 6 5.50 3.36 0.00 0.00 0.00 0.00 16 0.24 ?
QD1/2 LEU 6
HA LEU 6 5.22 4.44 0.00 0.00 0.63 0.00 12 0.38 ?
HG12/3 ILE 8
HA ILE 8 4.57 5.31 0.00 0.00 0.00 0.00 -10 0.00 ?
HB ILE 8 4.29 3.52 0.00 0.00 0.00 0.00 18 0.34 ?
HB2/3 LEU 9
H VAL 10 3.76 4.38 5.27 0.00 0.00 0.00 20 1.16 reversed
QD1/2 LEU 9
HA LEU 9 5.56 3.85 0.05 0.00 7.15 0.00 18 0.51 ?
QG1/2 VAL 10
H VAL 10 4.91 4.13 0.00 0.00 0.32 0.00 20 1.06 as input
HA VAL 10 3.70 4.72 0.00 0.00 1.10 0.00 -20 -0.90 as input
H CYS 11 4.97 5.87 0.00 0.00 0.00 0.00 -13 -0.10 ?
For every spin pair there are lines listing spins. NMR cross-peaks to these spins have been used in an attempt to determine stereospecific assignments.
-- Main.GaohuaLiu - 29 Jan 2007
- run_glomsa.cya: CYANA script to run GLOMSA