PSVS: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
== '''Introduction''' == | == '''Introduction''' == | ||
[http://www-nmr.cabm.rutgers.edu/PSVS/ PSVS] (Protein Structure Validation Server) systematically evaluates the quality of protein structures, and reports a large set of quality scores and constraint analyses | [http://www-nmr.cabm.rutgers.edu/PSVS/ PSVS] (Protein Structure Validation Server) systematically evaluates the quality of protein structures, and reports a large set of quality scores and constraint analyses [1]. PSVS defines a set of standard quality scores and constraint analyses that are reported for each structure. In addition to experimental constraints, this set encompasses a number of parameters evaluating different aspects of structure quality, including fold and packing, local residue separation, deviations of bond length and bond angles, backbone and side-chain torsion angle conformation, and atomic overlaps. Quality scores are reported based on calibration with a set of high-resolution X-ray crystal structures. A Z-score for the query structure, calculated for the score from each tool, provides an evaluation of the quality of the structure compared to this set of X-ray crystal structures. This server returns a concise validation report that includes a standard set of graphs and tables. <br> | ||
<br> | <br> | ||
| Line 45: | Line 45: | ||
== '''References''' == | == '''References''' == | ||
1. Bhattacharya, A., Tejero, R., and Montelione, G. T. (2007) Evaluating protein structures determined by structural genomics consortia. ''Proteins 66'', 778-795. | [http://www3.interscience.wiley.com/journal/114029977/abstract 1. Bhattacharya, A., Tejero, R., and Montelione, G. T. (2007) Evaluating protein structures determined by structural genomics consortia. ''Proteins 66'', 778-795.] | ||
<br> -- Main.GaohuaLiu - 05 Feb 2007 | <br> -- Main.GaohuaLiu - 05 Feb 2007 | ||
-- Updated by JimAramini - 31 Oct 2009 | -- Updated by JimAramini - 31 Oct 2009 | ||
Revision as of 20:29, 23 December 2009
Introduction
PSVS (Protein Structure Validation Server) systematically evaluates the quality of protein structures, and reports a large set of quality scores and constraint analyses [1]. PSVS defines a set of standard quality scores and constraint analyses that are reported for each structure. In addition to experimental constraints, this set encompasses a number of parameters evaluating different aspects of structure quality, including fold and packing, local residue separation, deviations of bond length and bond angles, backbone and side-chain torsion angle conformation, and atomic overlaps. Quality scores are reported based on calibration with a set of high-resolution X-ray crystal structures. A Z-score for the query structure, calculated for the score from each tool, provides an evaluation of the quality of the structure compared to this set of X-ray crystal structures. This server returns a concise validation report that includes a standard set of graphs and tables.
Running the PSVS server
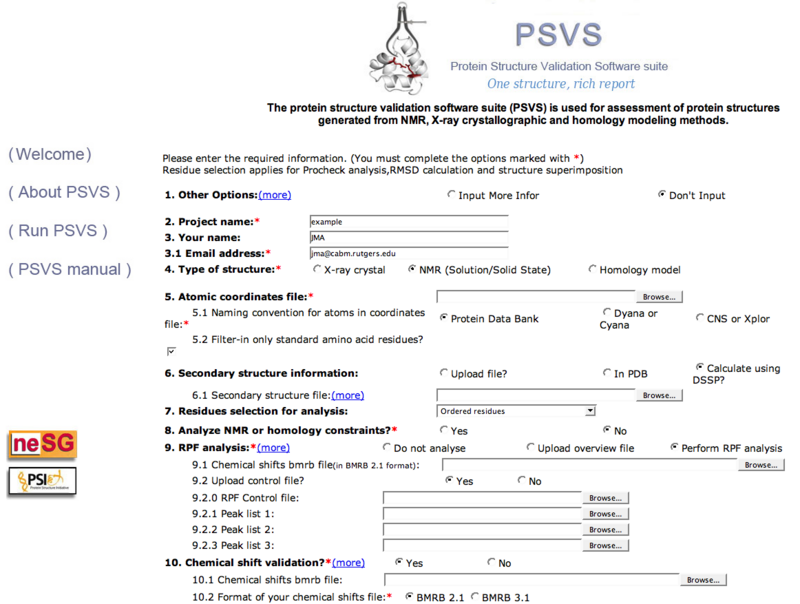
Once the structure is available, go to the PSVS server and upload the input files as described in that page (Figure 1).
- PDB coordinates: atom names can be in PDB format, DYANA/CYANA or CNS/XPLOR format
- Peak lists (optional, for RPF scores check): separate peak lists from 15N-resolved NOESY and 13C-resolved NOESY.
- Chemical shift (optional, for constraints violation and RPF scores check): BMRB 2.1 or 3.1 format
- Constraints (optional, for constraints violation check): can be in CYANA or XPLOR/CNS format.
Figure 1: PSVS input page
PSVS Output
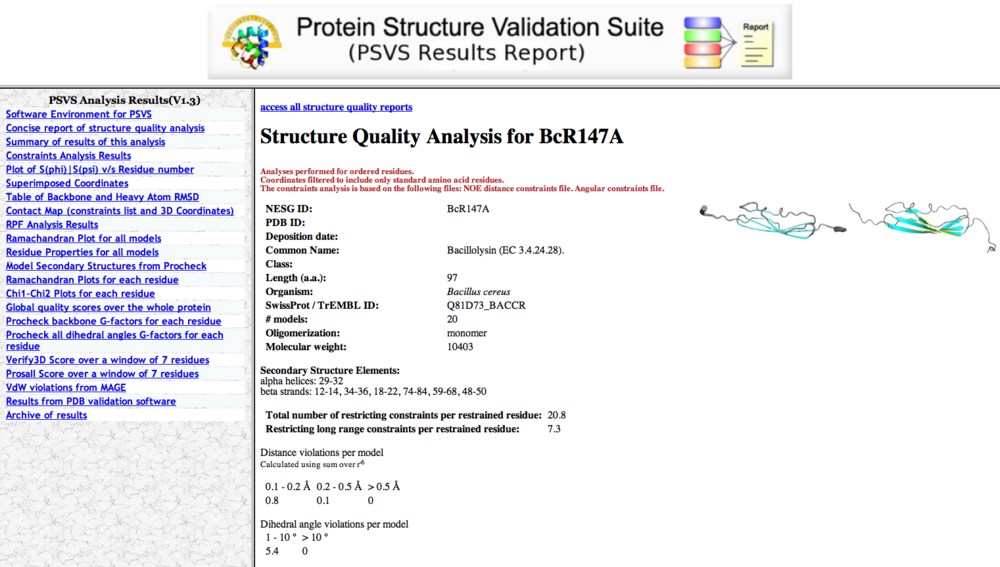
One will get an e-mail notice a few minutes after PSVS submisson, and following the link in the e-mail will lead to the PSVS results (Figure 2). Various outputs can be accessed from tabs in the left margin, and a zipped file of the entire analysis can be downloaded.
Typical PSVS analyses include:
- Secondary Structure Elements
- Ordered residues
- RMSD values for all residues and ordered residues.
- Ramachandran Plot Summary for ordered residues from Procheck
- Global quality scores for Verify3D, ProsaII, Procheck (phi-psi), Procheck (all) and MolProbity Clashscore. It is required that one should never deposit a structure to the PDB with Z scores < -5 for Procheck scores and Molprobity clashscores. General, Z szores > -3 for both Procheck G factor and Molprobity Clashscore are recommended.
- RPF Scores (optional). A good structure should have R > 90%, P > 90%, F> 90% and DP > 70%.
Figure 2: PSVS output page
References
-- Main.GaohuaLiu - 05 Feb 2007
-- Updated by JimAramini - 31 Oct 2009