REDCAT: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
== '''Introduction''' == | == '''Introduction''' == | ||
Recent availability of residual dipolar couplings (RDC) data has highlighted the need for general and user friendly RDC analysis tools. REDCAT (REsidual Dipolar Coupling Analysis Tool) has been designed and distributed as a general tool for analysis of RDC data (Ref. 1). REDCAT is a user-friendly program with its graphical-user-interface developed in Tcl/Tk and is therefore highly portable. The computational engine behind this GUI is written in C/C++ providing excellent computational performance. Separation of the computational engine from the graphical engine allows for flexible and easy command line interaction that can used for automated data analysis sessions. | Recent availability and utility of residual dipolar couplings (RDC) data has highlighted the need for general and user friendly RDC analysis tools. REDCAT (REsidual Dipolar Coupling Analysis Tool) has been designed and distributed as a general tool for analysis of RDC data (Ref. 1). REDCAT is a user-friendly program with its graphical-user-interface developed in Tcl/Tk and is therefore highly portable. The computational engine behind this GUI is written in C/C++ providing excellent computational performance. Separation of the computational engine from the graphical engine allows for flexible and easy command line interaction that can used for automated data analysis sessions. | ||
Conventional use of REDCAT requires existence of molecular structure and assigned RDC data. In the presence of structure and assigned data, REDCAT can be used to | Conventional use of REDCAT requires existence of molecular structure and assigned RDC data. In the presence of structure and assigned data, REDCAT can be used to scrutinize the validity of a structure, study internal motion, orient rigid domains with respect to each other and emulate RDC data under static and dynamic conditions. | ||
A number of improved algorithms have been incorporated to obtain order tensors and back calculate couplings from a given order tensor and proposed structure. These include the proper sampling of the Null-space (when the system of linear equations is underdetermined), more sophisticated filters for invalid order-tensor identification, error analysis for the identification of the problematic measurements and simulation of the effects of dynamic averaging process. | |||
More descriptive on-line documentation and tutorials on REDCAT can be found at the following [http://ifestos.cse.sc.edu/wiki/html/index.php/Redcat link]. | |||
== '''Features''' == | == '''Features''' == | ||
| Line 24: | Line 26: | ||
<br> | <br> | ||
== '''Downloading and Installing REDCAT''' == | |||
Instructions for downloading REDCAT and its installation can be found at this [http://ifestos.cse.sc.edu/wiki/html/index.php/Redcat:Installation link]. | |||
== '''Finding the Alignment Tensor Using REDCAT''' == | |||
*Prepare your rdc data as a single column text file with a list of rdc scalar couplings in hz. The rdc couplings must be from a block of consequtive residues within you protein. You must know the starting residue and ending residue numbers for this block. For residues within the block that you do not know the couplings for, use 999 to indicate an unknown coupling. | |||
*Prepare a pdb file with a single (best) model of your structure. | |||
*Start REDCAT for example by typing "REDCAT.tcl" | |||
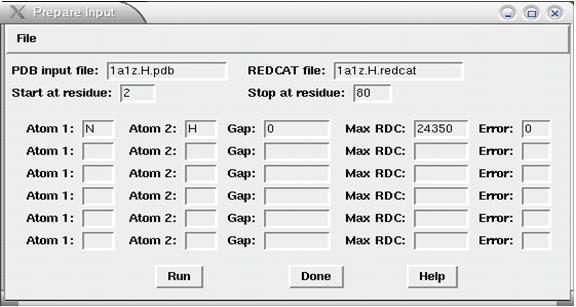
*Go to the 'FILE' menu and select 'Prepare Input' the menu will look like that shown in Figure 1.<br> | |||
==== Figure 1: Image of "prepare input" menu from REDCAT ==== | |||
[[Image:REDCATinputMenu.jpg]] | |||
*Enter the name of your PDB file under 'PDB Input file'. | |||
*Enter a name for the desired output REDCAT file. Example: myprot.redcat. | |||
*Enter the starting and ending resdue numbers for your block of RDC data (above). | |||
*Enter the atom types involved in the bond for which the RDC data was collected. These atom types must be consistent with the nomenclature in your pdb file. For example N, NH. | |||
*The Gap will usually be 0 indicating both atoms are from the same residue. | |||
*The Max RDC is the RDC value for this bond at perfect alignment observed at a 1 Angstrom difference. For NH it is 24350. For other values hit the HELP button. | |||
*The error should be put at about 10% of the spread in value of your RDC data (max RDC - min RDC). You may want to start with a smaller number and then relax it if necessary to incorporate more RDCs into your alignment tensor calculation. | |||
*Once you have input all these parameters, click on Done. | |||
*Now select the 'Load' command within the 'File' menu and load the 'myprot.redcat' file you made in the prepare input menu. | |||
*Select the 'Import RDC' command and enter the single column text file containing your RDC values. These will then populate your myprot.redcat file with the rdc values. RDC values of 999 will be automatically unselected in the REDCAT file. | |||
*Check that the rdc values are populating the correct resididues. | |||
*Now hit the 'run' button. | |||
*You should see a table showing each of the residues in either grey meaning not used, green meaning rdc accepted or red meaning rdc rejected. | |||
*Go through and unselect the red ones. Ideally, these are from loops or other disordered regions. If you have a lot of rejections from ordered regions, it means your structure does not agree well with the RDC values. | |||
*clear the output screen and 'run' again. Again, unselect any red RDCs. 'run' again. | |||
*When you get a run with no red rdc values, look at the top of the output, it may still say 10000 rejections out of 10000. If it does, look for the particular rdc values causing the most rejections. Unselect them. | |||
*Keep unselecting the worst rdc values until you get some solutions. | |||
*Go into 'tools', 'solutions', get solutions. | |||
*Write down the Sxx, Syy, and Szz values of the best solutions. These are relative to max rdc value. Multiply them by (for example 24350 for NH) to get the values to use for calculating Da and R. | |||
=== <br> === | |||
== '''Finding the Q-Factor Using REDCAT''' == | |||
*You must first have loaded or build a redcat file from a pdb structure, populated it with rdc values, and found the alignment tensor and euler angles as described above. | |||
*Once you have gotten a solution for the order parameters, go into the "Tools"/"Solutions"/"Get best Solution" in REDCAT menu and display (and or write down) the order parameters and euler angles. | |||
*Then: go into the "Tools"/"Calculations"/"Calculate/Substitute Rdc" menu. | |||
*Put in the Sxx, Syy, Szz values and the euler angles. You can round the Sxx, Syy and Szz parameters to a couple of significant digits, however Sxx + Syy + Szz must be exactly 0 or you will get an "invalid order parameters" message. | |||
*Put in an error of 0 | |||
*Hit the "Calculate Rdc" button. | |||
*The rmsd between the measured and calculated rdcs and the Quality Factor will show up at the bottom of the window. | |||
== '''References''' == | == '''References''' == | ||
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WJX-4BJWWY6-2&_user=521354&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000026038&_version=1&_urlVersion=0&_userid=521354&md5=978a5b8d1cf72110c78cc737e2e15195 1- Homayoun Valafar and James H. Prestegard, REDCAT: a residual dipolar coupling analysis tool, Journal of Magnetic Resonance, Volume 167, Issue 2, April 2004, Pages 228-241] <br> | [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WJX-4BJWWY6-2&_user=521354&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000026038&_version=1&_urlVersion=0&_userid=521354&md5=978a5b8d1cf72110c78cc737e2e15195 1- Homayoun Valafar and James H. Prestegard, REDCAT: a residual dipolar coupling analysis tool, Journal of Magnetic Resonance, Volume 167, Issue 2, April 2004, Pages 228-241] <br> | ||
-- SonalBansal - Sept 2007 | |||
-- HomayounValafar - Feb 2009 | |||
-- Edited by JimAramini - Dec 2009 | |||
<br> | <br> | ||
Revision as of 18:27, 2 December 2009
Introduction
Recent availability and utility of residual dipolar couplings (RDC) data has highlighted the need for general and user friendly RDC analysis tools. REDCAT (REsidual Dipolar Coupling Analysis Tool) has been designed and distributed as a general tool for analysis of RDC data (Ref. 1). REDCAT is a user-friendly program with its graphical-user-interface developed in Tcl/Tk and is therefore highly portable. The computational engine behind this GUI is written in C/C++ providing excellent computational performance. Separation of the computational engine from the graphical engine allows for flexible and easy command line interaction that can used for automated data analysis sessions.
Conventional use of REDCAT requires existence of molecular structure and assigned RDC data. In the presence of structure and assigned data, REDCAT can be used to scrutinize the validity of a structure, study internal motion, orient rigid domains with respect to each other and emulate RDC data under static and dynamic conditions.
A number of improved algorithms have been incorporated to obtain order tensors and back calculate couplings from a given order tensor and proposed structure. These include the proper sampling of the Null-space (when the system of linear equations is underdetermined), more sophisticated filters for invalid order-tensor identification, error analysis for the identification of the problematic measurements and simulation of the effects of dynamic averaging process.
More descriptive on-line documentation and tutorials on REDCAT can be found at the following link.
Features
The following lists some of the features of REDCAT:
- Easy manipulation of the data during an analysis session via the GUI implementation
- Easy extraction of principal order parameters, GDO, Szz, eta and the Euler angles that allow the transformation of a structure into its principle alignment frame (PAF)
- More meaningful methods of screening for valid order tensor solutions
- Back-calculation of RDCs with any given tensor and structure
- Calculation of the rmsd and Q-factor between back calculated and measured RDCs
- Report of the best solution tensor (in the rmsd sense)
- Error analysis that allows the identification and isolation of problematic measurements or structural errors
- Computation of error values that will produce solutions for the given coordinates and RDC data
- Dynamic averaging of RDCs due to motion
Downloading and Installing REDCAT
Instructions for downloading REDCAT and its installation can be found at this link.
Finding the Alignment Tensor Using REDCAT
- Prepare your rdc data as a single column text file with a list of rdc scalar couplings in hz. The rdc couplings must be from a block of consequtive residues within you protein. You must know the starting residue and ending residue numbers for this block. For residues within the block that you do not know the couplings for, use 999 to indicate an unknown coupling.
- Prepare a pdb file with a single (best) model of your structure.
- Start REDCAT for example by typing "REDCAT.tcl"
- Go to the 'FILE' menu and select 'Prepare Input' the menu will look like that shown in Figure 1.
- Enter the name of your PDB file under 'PDB Input file'.
- Enter a name for the desired output REDCAT file. Example: myprot.redcat.
- Enter the starting and ending resdue numbers for your block of RDC data (above).
- Enter the atom types involved in the bond for which the RDC data was collected. These atom types must be consistent with the nomenclature in your pdb file. For example N, NH.
- The Gap will usually be 0 indicating both atoms are from the same residue.
- The Max RDC is the RDC value for this bond at perfect alignment observed at a 1 Angstrom difference. For NH it is 24350. For other values hit the HELP button.
- The error should be put at about 10% of the spread in value of your RDC data (max RDC - min RDC). You may want to start with a smaller number and then relax it if necessary to incorporate more RDCs into your alignment tensor calculation.
- Once you have input all these parameters, click on Done.
- Now select the 'Load' command within the 'File' menu and load the 'myprot.redcat' file you made in the prepare input menu.
- Select the 'Import RDC' command and enter the single column text file containing your RDC values. These will then populate your myprot.redcat file with the rdc values. RDC values of 999 will be automatically unselected in the REDCAT file.
- Check that the rdc values are populating the correct resididues.
- Now hit the 'run' button.
- You should see a table showing each of the residues in either grey meaning not used, green meaning rdc accepted or red meaning rdc rejected.
- Go through and unselect the red ones. Ideally, these are from loops or other disordered regions. If you have a lot of rejections from ordered regions, it means your structure does not agree well with the RDC values.
- clear the output screen and 'run' again. Again, unselect any red RDCs. 'run' again.
- When you get a run with no red rdc values, look at the top of the output, it may still say 10000 rejections out of 10000. If it does, look for the particular rdc values causing the most rejections. Unselect them.
- Keep unselecting the worst rdc values until you get some solutions.
- Go into 'tools', 'solutions', get solutions.
- Write down the Sxx, Syy, and Szz values of the best solutions. These are relative to max rdc value. Multiply them by (for example 24350 for NH) to get the values to use for calculating Da and R.
Finding the Q-Factor Using REDCAT
- You must first have loaded or build a redcat file from a pdb structure, populated it with rdc values, and found the alignment tensor and euler angles as described above.
- Once you have gotten a solution for the order parameters, go into the "Tools"/"Solutions"/"Get best Solution" in REDCAT menu and display (and or write down) the order parameters and euler angles.
- Then: go into the "Tools"/"Calculations"/"Calculate/Substitute Rdc" menu.
- Put in the Sxx, Syy, Szz values and the euler angles. You can round the Sxx, Syy and Szz parameters to a couple of significant digits, however Sxx + Syy + Szz must be exactly 0 or you will get an "invalid order parameters" message.
- Put in an error of 0
- Hit the "Calculate Rdc" button.
- The rmsd between the measured and calculated rdcs and the Quality Factor will show up at the bottom of the window.
References
-- SonalBansal - Sept 2007
-- HomayounValafar - Feb 2009
-- Edited by JimAramini - Dec 2009