Resonance Assignment/Abacus/Spin systems identification: Difference between revisions
Jump to navigation
Jump to search
15N_HSQC
No edit summary |
RyanDoherty (talk | contribs) No edit summary |
||
| (2 intermediate revisions by one other user not shown) | |||
| Line 28: | Line 28: | ||
<div> </div><div><span> 2.2<span> </span></span>Read the generated peaks into SPARKY. </div><div> </div><div><span> 2.3<span> </span></span>Using SPARKY, complete ''b''PB fragments with aliphatic side-chain resonances by analyzing C<sub>a</sub>-H<sub>a</sub> and C<sub>b</sub>-H<sub>b</sub><span> strips in all CH_rooted spectra (see Figure 4.1B,C). New resonances should be peaked and referenced ''<u>only</u>'' in 13C_HSQC spectrum.</span></div><div> </div> | <div> </div><div><span> 2.2<span> </span></span>Read the generated peaks into SPARKY. </div><div> </div><div><span> 2.3<span> </span></span>Using SPARKY, complete ''b''PB fragments with aliphatic side-chain resonances by analyzing C<sub>a</sub>-H<sub>a</sub> and C<sub>b</sub>-H<sub>b</sub><span> strips in all CH_rooted spectra (see Figure 4.1B,C). New resonances should be peaked and referenced ''<u>only</u>'' in 13C_HSQC spectrum.</span></div><div> </div> | ||
=== Figure 4.1B,C === | === Figure 4.1B,C === | ||
<div></div><div></div><div>[[Image:Fmcgui Fig4.1b.jpg|thumb|left| | <div></div><div></div><div>[[Image:Fmcgui Fig4.1b.jpg|thumb|left|500px]][[Image:Fmcgui Fig4.1c.jpg|thumb|right|500px]]</div><div></div><div></div><div></div><div><span> </span></div><div><br></div><div><span>2.4<span> </span></span>When all peaks corresponding to HN-rooted spin-systems are peaked in 13C_HSQC spectrum, the unpicked peaks are used as a starting point for identification of spin-systems without backbone HN resonances (PB fragments corresponding to residues before prolines in the protein sequence, last residue and residues missing in HN-rooted spectra).</div><div> </div> | ||
<br> | <br> | ||
== Step 3. Spin-systems validation and correction == | == Step 3. Spin-systems validation and correction == | ||
<div> </div><div>Validate PB-fragments using FMCGUI:</div> | <div> </div><div>Validate PB-fragments using FMCGUI:</div> | ||
*<span> start a new project PRJ2 { [[FMCGUI commands#Project.3ENew|Project>new]] } | *<span> start a new project PRJ2 { [[FMCGUI commands#Project.3ENew|Project>new]] } </span> | ||
</span> | |||
<br> | <br> | ||
<br> | |||
*<span><span> </span></span>load protein sequence { [[FMCGUI commands#Data.3EProtein_Sequence.3ELoad|DATA>Protein Sequence>load]] } | *<span><span> </span></span>load protein sequence { [[FMCGUI commands#Data.3EProtein_Sequence.3ELoad|DATA>Protein Sequence>load]] } | ||
| Line 48: | Line 49: | ||
*<span><span> </span></span>set tolerances for matching of resonances in different spectral dimensions { [[FMCGUI commands#Data.3ETolerances|DATA>Tolerances]] } | *<span><span> </span></span>set tolerances for matching of resonances in different spectral dimensions { [[FMCGUI commands#Data.3ETolerances|DATA>Tolerances]] } | ||
*<span><span> </span></span>create and validate PB fragments { [[FMCGUI commands#Fragment.3ECreate.3Eabacus|Fragment>create>abacus]] } | *<span><span> </span></span>create and validate PB fragments { [[FMCGUI commands#Fragment.3ECreate.3Eabacus|Fragment>create>abacus]] } | ||
<div>This command starts script ‘''sps_create’''. In the result, a new window [[FMCGUI commands#Fragment.3ECreate.3Eabacus|"Create Fragment"]] pops up. The warning messages of the ‘''sps_create’'' script are shown in the main window and indicate spin-system that have low score (see [[Introduction to ABACUS#Figure_1.2|Figure 1.2]]), namely, spin | <div>This command starts script ‘''sps_create’''. In the result, a new window [[FMCGUI commands#Fragment.3ECreate.3Eabacus|"Create Fragment"]] pops up. The warning messages of the ‘''sps_create’'' script are shown in the main window and indicate spin-system that have low score (see [[Introduction to ABACUS#Figure_1.2|Figure 1.2]]), namely, spin-systems with S<sub>max</sub> < 10<sup>-4</sup>. Following the warnings check and modify, if necessary, generated PB fragments in the left section of “Create Fragment’ window. Alternatively, go back to spectra, fix peak lists accordingly, and repeat the fragment generation/validation again.</div><div>When satisfied, the PB fragments will be saved (file "sps_pb.dat" into directory "PRJ2/sps") and loaded in memory by pressing "OK" button in “Create Fragment” window.</div> | ||
*<span> save project PRJ2 { [[FMCGUI commands#Project.3ESave|Project>Save]] } or { [[FMCGUI commands#Project.3EQuit|Project>Quit]] } | *<span> save project PRJ2 { [[FMCGUI commands#Project.3ESave|Project>Save]] } or { [[FMCGUI commands#Project.3EQuit|Project>Quit]] } </span> | ||
</span> | |||
<div>''' '''</div> | |||
<div>''' '''</div> | |||
<br> | <br> | ||
Latest revision as of 21:18, 6 January 2010
Normally, the NMR spectra shown in Table 1 are collected for ABCUS. Spectra should ideally be collected from the same protein preparation and under the same conditions to make sure peaks are within tolerance between spectra. All spectra need to be appropriately synchronized and calibrated against reference spectra of your choice. It is a good idea to use as a reference for spectra calibration the 2D HN-projection of 15N-NOESY and the 2D CH-projection of the 13C-NOESY.
Step 1. Generate HN-rooted spin-systems (bPB-fragments)
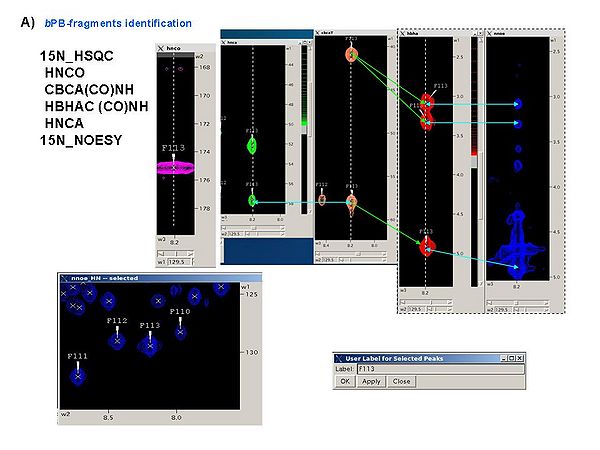
1.1bPB-fragments can be identified using the following spectra
15N_HSQC
HNCO
CBCA(CO)NH
HBHAC (CO)NH
HNCA
15N_NOESY
It is a good idea to analyze all these spectra simultaneously, for example, using SPARKY, in order to obtain peak lists of 15N_HSQC, HNCA, CBCA(CO)NH, and HBHA(CO)NH spectra (see Figure 4.1A). The first two spectra should be referenced.
- start new project PRJ_1 { Poject>new }
- load protein sequence { DATA>Protein Sequence>load }
- load referenced HNCA peak list { DATA>HNCA>load }
- load referenced CBCA(CO)NH peak list { DATA>CBCACONH>load }
- load referenced HBHA(CO)NH peak list { DATA>HBHACONH>load }
- load referenced 15N_HSQC peak list { DATA>N15 HSQC>load }
- set tolerances for matching of resonances in different spectral dimensions { Data>Tolerances }
- create bPB-fragments { Fragment>create>fawn }
First, a referenced C13_hsqc peak list is created and shown in new window ‘fake C13 HSQC’. Warning messages are shown in the project main window. You have to check the list and modify it if needed. Then, press ‘OK’ button in the ‘‘fake C13 HSQC’ window. In the result, new window ‘Create Fragment’ pops up. The window consists of three sections. The left sections contains suggested bPB-fragments, while the other sections contains two reports of fragments scoring with both C and H resonances and with only C resonances, respectively. Consider warning messages shown in the project main window and check/modify generated bPB-fragments in the left section of “Create Fragment” window. When satisfied, the bPB-fragments will be loaded in the memory by clicking on “OK” button.
- save project PRJ_1. { Project>Save } or { Project>Quit }
Step 2. Complete PB-fragments
At this step bPB-fragments are completed with aliphatic side-chain resonances and additional spin-systems (without HN) are identified using the following spectra.
13C-HSQC
(H)CCH-TOCSY
H(C)CH-TOCSY
13C-NOESY
2.1. Generate expected peak lists for C13HSQC, (H)CCH-TOCSY, and H(C)CH-TOCSY spectra using created bPB fragments:
- open project PRJ1 { Project>load }
- generate expected C13hsqc_exp.list that consists of Ca-Ha and Cb-Hb moieties { Fragment>Expected Peaks>C13HSQC }
- generate expected CCH-Tocsy.list { Fragment>Expected Peaks>(H)CCH }
- generate expected HCH-Tocsy.list { Fragment>Expected Peaks>H(C)CH }
2.2 Read the generated peaks into SPARKY.
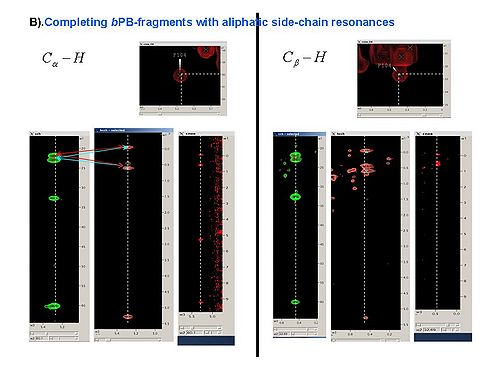
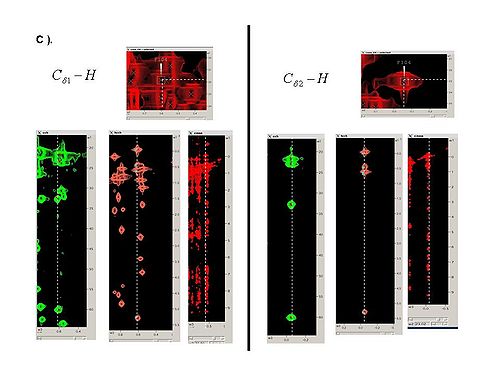
2.3 Using SPARKY, complete bPB fragments with aliphatic side-chain resonances by analyzing Ca-Ha and Cb-Hb strips in all CH_rooted spectra (see Figure 4.1B,C). New resonances should be peaked and referenced only in 13C_HSQC spectrum.
Figure 4.1B,C
2.4 When all peaks corresponding to HN-rooted spin-systems are peaked in 13C_HSQC spectrum, the unpicked peaks are used as a starting point for identification of spin-systems without backbone HN resonances (PB fragments corresponding to residues before prolines in the protein sequence, last residue and residues missing in HN-rooted spectra).
Step 3. Spin-systems validation and correction
Validate PB-fragments using FMCGUI:
- start a new project PRJ2 { Project>new }
- load protein sequence { DATA>Protein Sequence>load }
- load referenced 15N_HSQC peak list { DATA>N15 HSQC>load }
- load referenced 13C_HSQC peak list { DATA>C13 HSQC>load }
- load referenced HNCA peak list { DATA>HNCA>load }
- load CBCA(CO)HN peak list { DATA>CBCACOHN>load }
- load HNCO peak list { DATA>HNCO>load }
- set tolerances for matching of resonances in different spectral dimensions { DATA>Tolerances }
- create and validate PB fragments { Fragment>create>abacus }
This command starts script ‘sps_create’. In the result, a new window "Create Fragment" pops up. The warning messages of the ‘sps_create’ script are shown in the main window and indicate spin-system that have low score (see Figure 1.2), namely, spin-systems with Smax < 10-4. Following the warnings check and modify, if necessary, generated PB fragments in the left section of “Create Fragment’ window. Alternatively, go back to spectra, fix peak lists accordingly, and repeat the fragment generation/validation again.
When satisfied, the PB fragments will be saved (file "sps_pb.dat" into directory "PRJ2/sps") and loaded in memory by pressing "OK" button in “Create Fragment” window.
- save project PRJ2 { Project>Save } or { Project>Quit }