Resonance Assignment/AutoAssign: Difference between revisions
(Created page with '== '''Introduction''' == [http://www-nmr.cabm.rutgers.edu/NMRsoftware/nmr_software.html AutoAssign] is a constraint-based expert system for automating the assignment and analys…') |
No edit summary |
||
| Line 5: | Line 5: | ||
The following description is mainly taken from the [http://www-nmr.cabm.rutgers.edu/NMRsoftware/autoassign/Documentation/Help/tutorial.html AutoAssign SOP (tutorial)]. Please check this tutorial and the version 2.4.0 [http://www-nmr.cabm.rutgers.edu/NMRsoftware/autoassign/Documentation/index.html help documentation] for more information and usage. <br> | The following description is mainly taken from the [http://www-nmr.cabm.rutgers.edu/NMRsoftware/autoassign/Documentation/Help/tutorial.html AutoAssign SOP (tutorial)]. Please check this tutorial and the version 2.4.0 [http://www-nmr.cabm.rutgers.edu/NMRsoftware/autoassign/Documentation/index.html help documentation] for more information and usage. <br> | ||
== ''' | == '''Using AutoAssign''' == | ||
=== Input Files === | === Input Files === | ||
| Line 12: | Line 12: | ||
*A [[Media:PfR193An500s.fasta|sequence file]] in fasta format | *A [[Media:PfR193An500s.fasta|sequence file]] in fasta format | ||
*Peak lists for backbone experiments in Sparky format. | *Peak lists for backbone experiments in [[Sparky|Sparky]] format. | ||
=== Opening AutoAssign === | === Opening AutoAssign === | ||
| Line 25: | Line 25: | ||
[[Image:Autoassign1.jpg]] | [[Image:Autoassign1.jpg]] | ||
== '''Creating a Data Set'''<br> == | === '''Creating a Data Set'''<br> === | ||
=== Using the Peaklist Converter in the Graphical User Interface === | ==== Using the Peaklist Converter in the Graphical User Interface ==== | ||
The easiest way to create a dataset is to using the Dataset Creation Graphical User Interface. This interface will guide you through the basic steps of converting your peak lists, registering your peak lists, creating a control file, and evaluating the quality of your peak lists. Go to <code>File</code> -> <code>Convert/Create Dataset</code> from the main menu option to start the Dataset Creation Graphical User Interface (Figure 2).<br> | |||
[[Image:Autoassign2.jpg]]<br> | |||
== '''References''' == | == '''References''' == | ||
1. Zimmerman, D.E., Kulikowski, C.A., Huang, Y., Feng, W., Tashiro, M., Shimotakahara, S., Chien, C., Powers, R. and Montelione, G.T. (1997) Automated analysis of protein NMR assignments using methods from artificial intelligence. ''J. Mol. Biol. 269'', 592-610. <br> | 1. Zimmerman, D.E., Kulikowski, C.A., Huang, Y., Feng, W., Tashiro, M., Shimotakahara, S., Chien, C., Powers, R. and Montelione, G.T. (1997) Automated analysis of protein NMR assignments using methods from artificial intelligence. ''J. Mol. Biol. 269'', 592-610. <br> | ||
Revision as of 18:45, 9 November 2009
Introduction
AutoAssign is a constraint-based expert system for automating the assignment and analysis of backbone NMR resonance assignments of proteins (Ref. 1). The program is implemented in C++, Java2, and Perl programming languages and supported on SGI-IRIX, Sun-Solaris, MAC-OSX, x86-Linux, and x86_64-Linux architectures. The newest AutoAssign distribution (version 2.4.0) automates the assignments of HN, NH, CO, CA, CB, HA, and HB resonances in non-, partially-, and fully-deuterated samples. The rich graphical user interface (GUI) provides a many sets of tools for dataset conversions, assignment validations, and various graphical displays of assignment results. AutoAssign is well tested on a large number of independently-collected triple-resonance NMR data sets of proteins ranging in size from ~6 to ~32 kD, including one fully-deuterated protein and and a dataset with reduced-dimensionality experiments. AutoAssign performs the automated analysis of assignments in only seconds on current RISC and x86 platforms.
The following description is mainly taken from the AutoAssign SOP (tutorial). Please check this tutorial and the version 2.4.0 help documentation for more information and usage.
Using AutoAssign
Input Files
The input files for AutoAssign are:
- A sequence file in fasta format
- Peak lists for backbone experiments in Sparky format.
Opening AutoAssign
- Run the startup script
autoassignIf you are running the client on the same machine as the server. - If you are running the client on a machine different from the server, then you must first make sure that the server is running on the other machine. The easiest way to do this is to run the startup script
autoserveron this other computer.
The main AutoAssign window will appear (Figure 1).
Figure 1: AutoAssign Main Window
Creating a Data Set
Using the Peaklist Converter in the Graphical User Interface
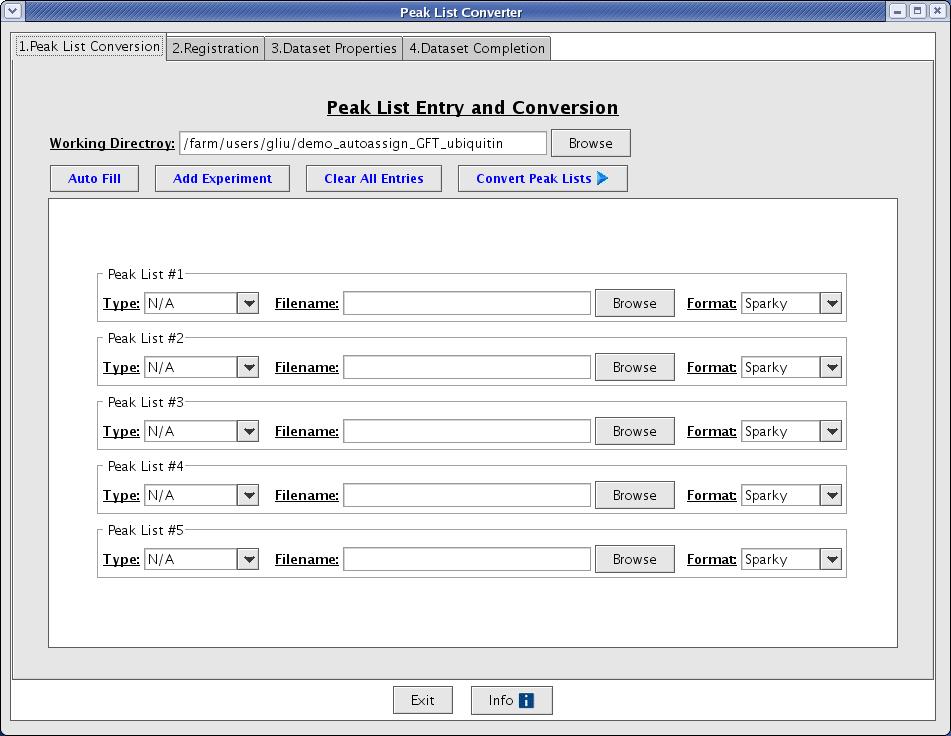
The easiest way to create a dataset is to using the Dataset Creation Graphical User Interface. This interface will guide you through the basic steps of converting your peak lists, registering your peak lists, creating a control file, and evaluating the quality of your peak lists. Go to File -> Convert/Create Dataset from the main menu option to start the Dataset Creation Graphical User Interface (Figure 2).
References
1. Zimmerman, D.E., Kulikowski, C.A., Huang, Y., Feng, W., Tashiro, M., Shimotakahara, S., Chien, C., Powers, R. and Montelione, G.T. (1997) Automated analysis of protein NMR assignments using methods from artificial intelligence. J. Mol. Biol. 269, 592-610.