Resonance Assignment/The PINE Server
Introduction
The recently developed PINE program (Probablistic Interaction Network of Evidence) uses a novel aglorithm for the automated assignment of both backbone and side chain protein NMR resonances (Ref. 1). PINE is available from a freely accessible web server. It is reported to have > 90% accuracy for backbone resonances and > 80% for aliphatic side chain resonances.
Using the PINE Server
Input Files
- amino acid sequence of the protein target either in single-letter or three-letter format.
- spectral peak lists. The program can accept peak lists in several formats including:
- SPARKY
- XEASY
- PINE
- additional information including:
- pre-assigned resonances
- amino acid selective labelling information
- grouped spins systems
The PINE Server
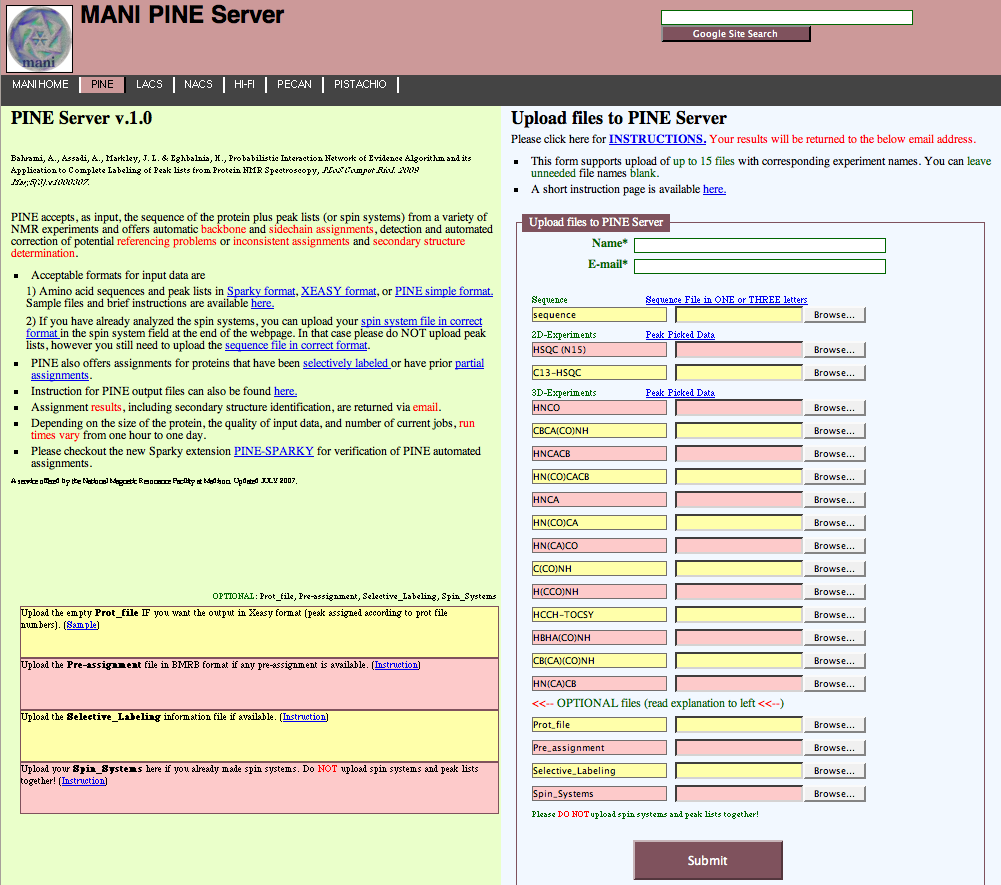
The PINE server is relatively simple to use (Figure 1). The user uploads the protein sequence, spectral peak lists and any additional information and simply submits to the server. There are extensive help links for the user on this main page. Note that for Sparjy peak lists, the current version of the server (1.0) cannot tolerate the Notes column; so, remove this column prior to submission.
Figure 1: The PINE Server
===
PINE Output
References
1. Bahrami, A., Assadi, A.H., Markley, J.L. and Eghbalnia, H.R. (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 5, e1000307.