XEASY Spin system identification: Difference between revisions
(Created page with ' == '''Spin System Identification''' == 2D [15N,1H]-HSQC provide pairs of correlated amide 15N / 1HN chemical shifts. They seed '''spin systems''' - spins of individual r…') |
No edit summary |
||
| (22 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<br> | |||
<br> | |||
== Spin System Identification == | |||
2D [15N,1H]-HSQC provide pairs of correlated amide 15N / 1HN chemical shifts. They seed '''spin systems''' - spins of individual residues, whose assignment to the protein sequence is normally not known ''a priori''. In an [[XEASY|'''XEASY''']]-based approach, spin systems are called '''SRD''' spin systems. | |||
(3,2)D HNNCO or 3D HNNCO provide additional resolution when both 15N and 1HN chemical shifts overlap, and help exclude side-chain peaks. Although 13C' chemical shifts are seldom used for sequence-specific assignment, they are used by the programs [[CSI|'''CSI''']] and [[TALOS|'''TALOS''']]. | |||
=== '''Spin System Identification with XEASY/UBNMR''' === | |||
( | ==== '''Peak Picking the (15N,1H) HSQC Spectrum''' ==== | ||
#Go to <tt>/protName/analysis/xeasy/nhsqc.</tt> Create a file with the amino acid sequence in [[Fasta|FASTA]] format, obtained, for example, from the [http://spine.nesg.org/ SPINE] web page, and save it as <tt>aa.seq</tt>. Run the <tt>makeSeq</tt> macro with [[UBNMR|UBNMR]] to generate an [[XEASY|XEASY]] [[XEASY Sequence List|SequenceList]] as <tt>nhsqc.seq</tt> . This [[XEASY Sequence List|SequenceList]] contains entries for the amino acids residues first, followed by two sets of '''SRD'''s named in the following way: '''SRD-I''' (Starting from 201) and '''SRD-II''' (starting from 401). '''SRD-II''' entires serve to handle sequential connectivities. See [[XEASY|XEASY]] Files for book-keeping: [[XEASY Atom List|AtomList]], [[XEASY Sequence List|SequenceList]], and [[XEASY Peak List|PeakList]]. | |||
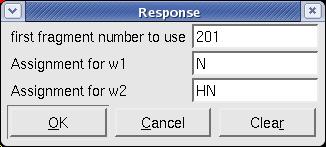
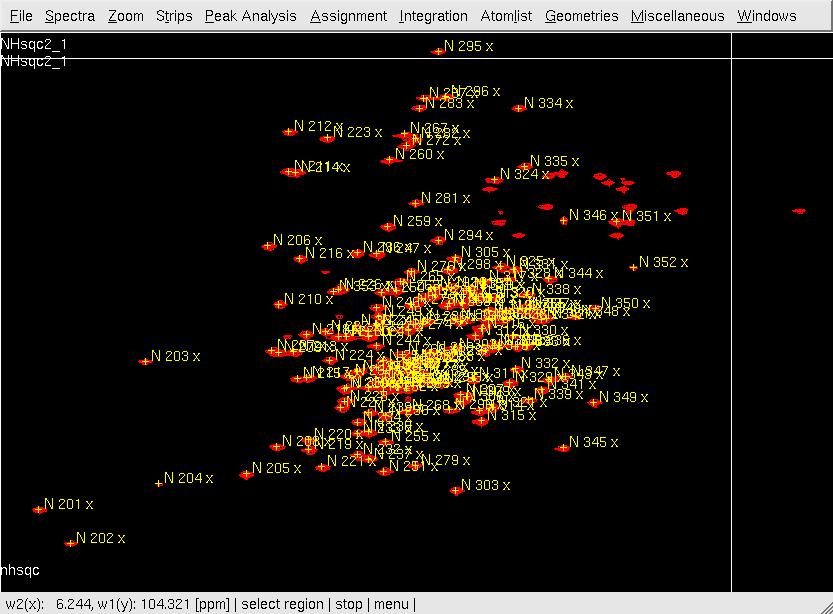
#In XEASY, use <tt>ns</tt>, <tt>ll</tt> and <tt>ls</tt> to load 2D [15N,1H]-HSQC spectrum, [[XEASY|XEASY]] library and [[XEASY Sequence List|SequenceList]]. To change display from the default one to contour plot, type <tt>cp</tt> and give the appropriate threshold level. A corresponding initial [[XEASY Atom List|AtomList]] is generated automatically. Use <tt>in</tt> for automatic in-phase peak picking of the 2D [15N,1H]-HSQC spectrum. Complete peak picking manually by using <tt>dp</tt> to remove the peaks belonging to sidechain amides which can be identified by a NH2only HSQC if the region is crowded and <tt>pp</tt> to pick additional peaks (Figure.1A). Use <tt>ar</tt> to automatically assign each peak to the backbone amide moiety of '''SRD-I''' residues (Figure.1B,C). Occasionally, the <tt>ar</tt> command causes an error <tt>Atom N 201 not known!</tt> which can be solved by loading the library file [<tt>ll</tt>] from <tt>/nsm/chem/cen2/HTP2/3_src/xeasy/src.new/xeasy.lib</tt> and then repeating the <tt>ar</tt> command. Average the chemical shifts and save the [[XEASY Peak List|PeakList]] (<tt>nhsqcO1.peaks</tt>) and [[XEASY Atom List|AtomList]] (<tt>nhsqcO1.prot</tt>) using <tt>ac</tt>, <tt>wc</tt> and <tt>wp</tt>. Then, amide 15N / 1HN chemical shifts are transferred into the [[XEASY Atom List|AtomList]] entries corresponding to '''SRD-I'''. | |||
<br><br>'''Figure 1.''' Peak picking the 2D [15N, 1H]-HSQC spectrum | |||
'''A: After in-phase peak picking'''<br> [[Image:XEASY hsqc2.jpg]] <br> <br> | |||
</ | '''B: XEASY <tt>ar</tt> window'''<br> [[Image:XEASY hsqc ar.jpg]]<br> | ||
''' | <br> '''C: After assigning the peaks to SRD-I numbers using <tt>ar</tt>'''<br> | ||
[[Image:XEASY hsqc3.jpg]]<br> | |||
< | |||
<br> | |||
==== '''Analysis of the 3D HNNCO Spectrum''' ==== | |||
#Go to <tt>/protName/analysis/xeasy/hnco</tt> and make a copy of HNCO spectrum (<tt>cp HNCO1.3D.16 </tt><tt>HNCO1a.3D.16</tt> and <tt>cp HNCO1.3D.param </tt><tt>HNCO1a.3D.param</tt>). | |||
#Run the <tt>makeHncoPeak</tt> script (see below) in [[UBNMR|UBNMR]].This [[XEASY Peak List|PeakList]] will contain one peak for each amide N(i) / H(i) moiety which will be assigned to an '''SRD-I''' number derived from the 2D [15N,1H]-HSQC [[XEASY Peak List|PeakList]]. The third / 13C'(i-1) dimension is assigned to the corresponding '''SRD-II''' number and has a default chemical shift of 175ppm. | |||
#Start [[XEASY|XEASY]] and use <tt>ns</tt> to load both HNNCO spectra <tt>HNCO1</tt> and <tt>HNCO1a</tt> with permutations set to <tt>x:HN, y:C, Z:N</tt> and <tt>x:N, y:C, Z:HN</tt>, respectively. Use <tt>ls</tt> to load the [[XEASY Sequence List|SeqList]] (<tt>nhsqc.seq</tt>), <tt>lc</tt> to load the [[XEASY Atom List|AtomList]] (<tt>nhsqcO1.prot</tt>), <tt>lp</tt> to load the [[XEASY Peak List|PeakList]] (<tt>hncoI1.peak</tt>).<br> | |||
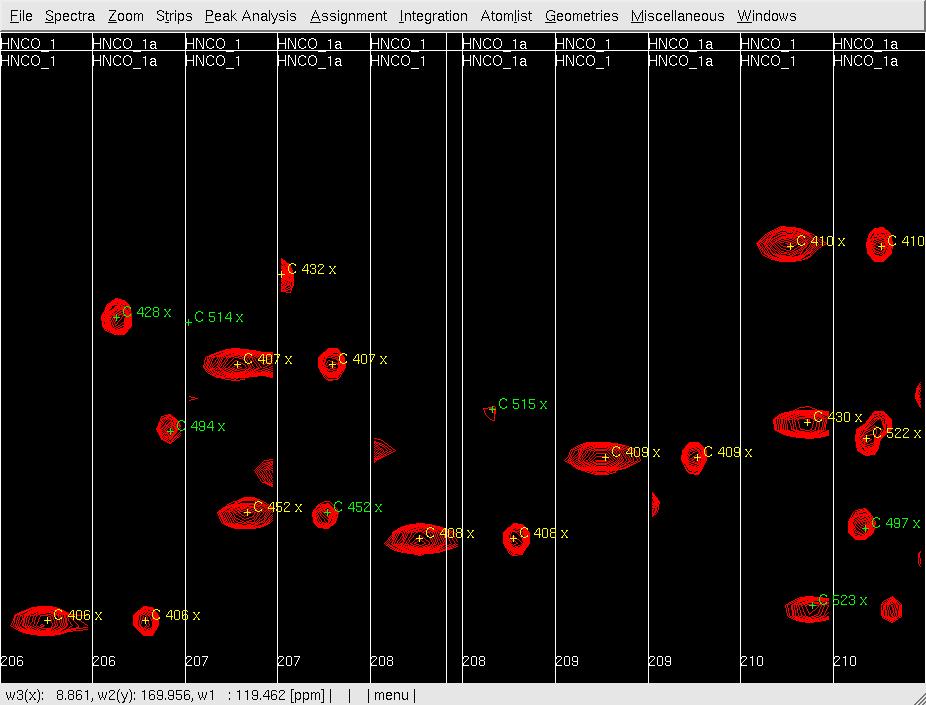
#Use <tt>cp</tt> to display the spectrum as a contour plot. Next, create the strips using <tt>se</tt> in the <tt>HNCO1</tt> spectrum, then change to <tt>HNCO1a</tt> using <tt>rc</tt> and <tt>rs</tt>, and create strips using <tt>se</tt> again in the <tt>HNCO1 </tt>spectrum. Use <tt>gs</tt> to display the strips (Figure 2A). | |||
#Use <tt>mr</tt> to move each pre-positioned peak onto the actual peak (Figure 2B). | |||
#Use <tt>dp</tt> to delete predicted peak markers that do not correspond to actual peaks in the spectrum (side-chain peaks) and <tt>pp</tt> to manually pick and assign (<span style="font-family: monospace;">a</span><tt>p</tt>) newly observed peaks (usually a result of overlap in the 2D [15N, 1H]-HSQC). | |||
#After all peaks are 'picked', use <tt>ac</tt> to update the chemical shifts, <tt>wc</tt> to save the [[XEASY Atom List|AtomList]] as <tt>hncoO1.prot</tt>, and <tt>wp</tt> to save the [[XEASY Peak List|PeakList]] as <tt>hncoO1.peaks</tt>. | |||
#Go to next step for [[XEASY Backbone Assignment|backbone assignment]]. | |||
<br> | |||
An example of the <tt>makeHncoPeak</tt> [[UBNMR|UBNMR]] script | |||
<pre>init | |||
read seq nhsqcO1.seq | |||
read prot nhsqcO1.prot | |||
write prot hncoI1.prot | |||
simulate 3D N C -1 2 | |||
write peaks hncoI1.peaks | |||
</pre> | |||
<br> | |||
<br>'''Figure 2: Analysis of the 3D HNNCO by XEASY''' <br>'''A: Before <tt>mr</tt>, showing orthogonal views of each strip'''<br>[[Image:XEASY hnco1.jpg]] | |||
<br>'''B: After <tt>mr</tt>'''<br>[[Image:XEASY hnco2.jpg]] <br> | |||
==== | |||
< | |||
==== '''Analysis of the (3,2)D GFT HNNCO spectrum''' ==== | |||
#Go to the <tt>/protName/analysis/xeasy/hnco</tt> directory.<br> | |||
#Run <tt>make32DHncoPeak</tt> in [[UBNMR|UBNMR]] (see below) to create a (3,2)D GFT HNNCO peaklist. | |||
< | #Follow the protocol shown above for assignment of 3D HNNCO spectra.<br> | ||
< | |||
< | |||
< | |||
<br> | |||
-- Main.GaohuaLiu - 16 Feb 2007 | An example of the <tt>make32DHncoPeak</tt> [[UBNMR|UBNMR]] script | ||
<pre>init | |||
read seq nhsqcO1.seq | |||
read prot nhsqcO1.prot | |||
write prot hncoI1.prot | |||
simulate 2D N+pC HN | |||
simulate 2D N-pC HN | |||
write peaks hncoI1.peaks | |||
</pre> | |||
<br> -- Main.GaohuaLiu - 16 Feb 2007<br> | |||
Latest revision as of 18:33, 24 November 2009
Spin System Identification
2D [15N,1H]-HSQC provide pairs of correlated amide 15N / 1HN chemical shifts. They seed spin systems - spins of individual residues, whose assignment to the protein sequence is normally not known a priori. In an XEASY-based approach, spin systems are called SRD spin systems.
(3,2)D HNNCO or 3D HNNCO provide additional resolution when both 15N and 1HN chemical shifts overlap, and help exclude side-chain peaks. Although 13C' chemical shifts are seldom used for sequence-specific assignment, they are used by the programs CSI and TALOS.
Spin System Identification with XEASY/UBNMR
Peak Picking the (15N,1H) HSQC Spectrum
- Go to /protName/analysis/xeasy/nhsqc. Create a file with the amino acid sequence in FASTA format, obtained, for example, from the SPINE web page, and save it as aa.seq. Run the makeSeq macro with UBNMR to generate an XEASY SequenceList as nhsqc.seq . This SequenceList contains entries for the amino acids residues first, followed by two sets of SRDs named in the following way: SRD-I (Starting from 201) and SRD-II (starting from 401). SRD-II entires serve to handle sequential connectivities. See XEASY Files for book-keeping: AtomList, SequenceList, and PeakList.
- In XEASY, use ns, ll and ls to load 2D [15N,1H]-HSQC spectrum, XEASY library and SequenceList. To change display from the default one to contour plot, type cp and give the appropriate threshold level. A corresponding initial AtomList is generated automatically. Use in for automatic in-phase peak picking of the 2D [15N,1H]-HSQC spectrum. Complete peak picking manually by using dp to remove the peaks belonging to sidechain amides which can be identified by a NH2only HSQC if the region is crowded and pp to pick additional peaks (Figure.1A). Use ar to automatically assign each peak to the backbone amide moiety of SRD-I residues (Figure.1B,C). Occasionally, the ar command causes an error Atom N 201 not known! which can be solved by loading the library file [ll] from /nsm/chem/cen2/HTP2/3_src/xeasy/src.new/xeasy.lib and then repeating the ar command. Average the chemical shifts and save the PeakList (nhsqcO1.peaks) and AtomList (nhsqcO1.prot) using ac, wc and wp. Then, amide 15N / 1HN chemical shifts are transferred into the AtomList entries corresponding to SRD-I.
Figure 1. Peak picking the 2D [15N, 1H]-HSQC spectrum
A: After in-phase peak picking
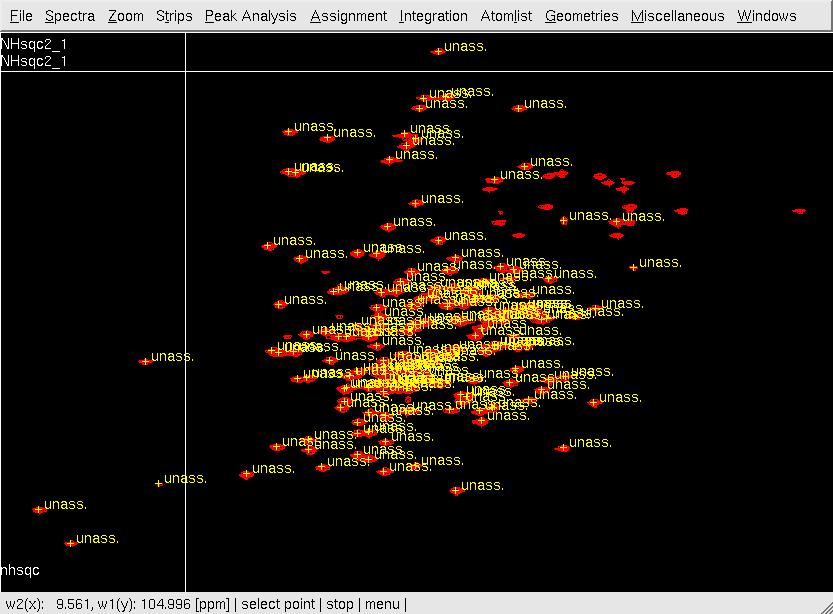
C: After assigning the peaks to SRD-I numbers using ar
Analysis of the 3D HNNCO Spectrum
- Go to /protName/analysis/xeasy/hnco and make a copy of HNCO spectrum (cp HNCO1.3D.16 HNCO1a.3D.16 and cp HNCO1.3D.param HNCO1a.3D.param).
- Run the makeHncoPeak script (see below) in UBNMR.This PeakList will contain one peak for each amide N(i) / H(i) moiety which will be assigned to an SRD-I number derived from the 2D [15N,1H]-HSQC PeakList. The third / 13C'(i-1) dimension is assigned to the corresponding SRD-II number and has a default chemical shift of 175ppm.
- Start XEASY and use ns to load both HNNCO spectra HNCO1 and HNCO1a with permutations set to x:HN, y:C, Z:N and x:N, y:C, Z:HN, respectively. Use ls to load the SeqList (nhsqc.seq), lc to load the AtomList (nhsqcO1.prot), lp to load the PeakList (hncoI1.peak).
- Use cp to display the spectrum as a contour plot. Next, create the strips using se in the HNCO1 spectrum, then change to HNCO1a using rc and rs, and create strips using se again in the HNCO1 spectrum. Use gs to display the strips (Figure 2A).
- Use mr to move each pre-positioned peak onto the actual peak (Figure 2B).
- Use dp to delete predicted peak markers that do not correspond to actual peaks in the spectrum (side-chain peaks) and pp to manually pick and assign (ap) newly observed peaks (usually a result of overlap in the 2D [15N, 1H]-HSQC).
- After all peaks are 'picked', use ac to update the chemical shifts, wc to save the AtomList as hncoO1.prot, and wp to save the PeakList as hncoO1.peaks.
- Go to next step for backbone assignment.
An example of the makeHncoPeak UBNMR script
init read seq nhsqcO1.seq read prot nhsqcO1.prot write prot hncoI1.prot simulate 3D N C -1 2 write peaks hncoI1.peaks
Figure 2: Analysis of the 3D HNNCO by XEASY
A: Before mr, showing orthogonal views of each strip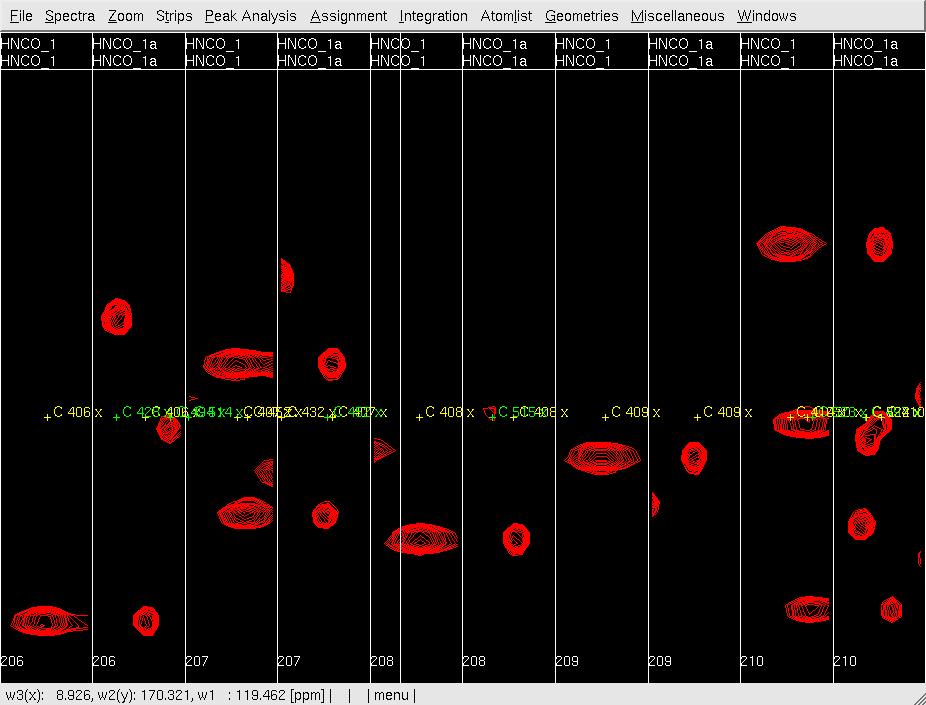
====
Analysis of the (3,2)D GFT HNNCO spectrum
- Go to the /protName/analysis/xeasy/hnco directory.
- Run make32DHncoPeak in UBNMR (see below) to create a (3,2)D GFT HNNCO peaklist.
- Follow the protocol shown above for assignment of 3D HNNCO spectra.
An example of the make32DHncoPeak UBNMR script
init read seq nhsqcO1.seq read prot nhsqcO1.prot write prot hncoI1.prot simulate 2D N+pC HN simulate 2D N-pC HN write peaks hncoI1.peaks
-- Main.GaohuaLiu - 16 Feb 2007