Resonance Assignment/Principles and concepts: Difference between revisions
mNo edit summary |
No edit summary |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 38: | Line 38: | ||
*(HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE, 2D - correlate H<span class="texhtml">δ</span> and H<span class="texhtml">ε</span> aromatic protons with C<span class="texhtml">β </span>of the same residue. Useful for assigning chemical shifts of nuclei in aromatic rings.<span class="texhtml"> | *(HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE, 2D - correlate H<span class="texhtml">δ</span> and H<span class="texhtml">ε</span> aromatic protons with C<span class="texhtml">β </span>of the same residue. Useful for assigning chemical shifts of nuclei in aromatic rings.<span class="texhtml"> | ||
</span> | </span> | ||
* | |||
*Long range <sup>15</sup>N-HSQC, 2D - correlates H<span class="texhtml">δ</span>2 and H<span class="texhtml">ε</span>1 protons (attached to <sup>12</sup>C or <sup>13</sup>C nuclei!) on His rings with N<span class="texhtml">δ1 </span>and N<span class="texhtml">ε2 </span>nuclei. This experiment s useful for assigning H<span class="texhtml">ε</span>1 and C<span class="texhtml">ε</span>1 nuclei of His rings as well as to determine the protonation state of the ring. | *Long range <sup>15</sup>N-HSQC, 2D - correlates H<span class="texhtml">δ</span>2 and H<span class="texhtml">ε</span>1 protons (attached to <sup>12</sup>C or <sup>13</sup>C nuclei!) on His rings with N<span class="texhtml">δ1 </span>and N<span class="texhtml">ε2 </span>nuclei. This experiment s useful for assigning H<span class="texhtml">ε</span>1 and C<span class="texhtml">ε</span>1 nuclei of His rings as well as to determine the protonation state of the ring. | ||
| Line 51: | Line 46: | ||
Through-space experiments, as the name implies, correlate pairs of nuclei close in space. Many such nuclei would belong to the same or adjacent residues and these experiments can thus supplement the information obtained from through-bond spectra. The usual distance limit to observe such a correlation is ~5Â. | Through-space experiments, as the name implies, correlate pairs of nuclei close in space. Many such nuclei would belong to the same or adjacent residues and these experiments can thus supplement the information obtained from through-bond spectra. The usual distance limit to observe such a correlation is ~5Â. | ||
*<sup>15</sup>N-edited NOESY-HSQC, 3D - correlates all protons close to the <sup>1</sup>H of an amide group with both nuclei of the amide | *<sup>15</sup>N-edited NOESY-HSQC, 3D - correlates all protons close to the <sup>1</sup>H of an amide group with both nuclei of the amide. | ||
*<sup>13</sup>C-edited NOESY-HSQC, 3D - correlates all protons close to the <sup>1</sup>H of a C-H moiety with both nuclei of the moiety. Can replace an H(C)CH-TOCSY experiment in certain cases. | *<sup>13</sup>C-edited NOESY-HSQC, 3D - correlates all protons close to the <sup>1</sup>H of a C-H moiety with both nuclei of the moiety. Can replace an H(C)CH-TOCSY experiment in certain cases. | ||
The above spectrum can be separated into aliphatic and aromatic regions of the <sup>13</sup>C range. | |||
*Simultaneous <sup>13</sup>C,<sup>15</sup>N-edited NOESY-HSQC, 3D - as the name implies, this spectrum combines the above two experiments (<sup>15</sup>N and <sup>13</sup>C). | *Simultaneous <sup>13</sup>C,<sup>15</sup>N-edited NOESY-HSQC, 3D - as the name implies, this spectrum combines the above two experiments (<sup>15</sup>N and <sup>13</sup>C). | ||
These NOESY-type spectra are very useful in confirming the sequence specific backbone assignments and in connecting aromatic rings to their proper side chains. For certain automatic methods (e.g. [[Abacus|ABACUS]]) they | These NOESY-type spectra are very useful in confirming the sequence-specific backbone assignments and in connecting aromatic rings to their proper side chains. For certain automatic methods (e.g. [[Abacus|ABACUS]]) they are used to establish sequential connectivities so that they can completely replace HNCACB or similar spectra. | ||
== Spin systems == | == Spin systems == | ||
| Line 66: | Line 63: | ||
Sometimes it may be more convenient to define a '''peptide-bond fragment''' (PB-fragment) spin system, which would contain N and HN of the following amino acid, instead of the same. See figure (left).<br> | Sometimes it may be more convenient to define a '''peptide-bond fragment''' (PB-fragment) spin system, which would contain N and HN of the following amino acid, instead of the same. See figure (left).<br> | ||
Occasionally, an aromatic ring may be treated as a separate spin system from the rest of the same side chain. | |||
=== [[Image:SpinSystemID.jpg|thumb|left|400px]]Identification === | === [[Image:SpinSystemID.jpg|thumb|left|400px]]Identification === | ||
| Line 76: | Line 73: | ||
=== Linking spin systems === | === Linking spin systems === | ||
Once identified, the spin systems can then be linked using information from relevant pairs of spectra, such as HNCACB and CBCA(CO)NH, HN(CO)CA and HNCA, HNCO and HN(CA)CO. Each software package has a slightly different method to do that, but generally for manual linking one looks for identical 13C chemical shifts in intraresidual and sequential spectra, simultaenously displaying strips from the above pairs. The programs often | Once identified, the spin systems can then be linked using information from relevant pairs of spectra, such as HNCACB and CBCA(CO)NH, HN(CO)CA and HNCA, HNCO and HN(CA)CO. Each software package has a slightly different method to do that, but generally for manual linking one looks for identical 13C chemical shifts in intraresidual and sequential spectra, simultaenously displaying strips from the above pairs. The programs often suggest the best fit. Some programs have built-in optimization functions.<br> | ||
Many automatic programs (MONTE, MARS, [[The PINE Server|PINE]], [[AutoAssign|AutoAssign]]) only require peak lists from the above spectra (and <sup>15</sup>N-HSQC) and use sofisticated algorithms to link the spin systems sequentially and match them onto the protein sequence. | Many automatic programs (MONTE, MARS, [[The PINE Server|PINE]], [[AutoAssign|AutoAssign]]) only require peak lists from the above spectra (and <sup>15</sup>N-HSQC) and use sofisticated algorithms to link the spin systems sequentially and match them onto the protein sequence. | ||
=== Matching onto covalent structure === | === Matching onto covalent structure === | ||
| Line 84: | Line 81: | ||
Some amino acids have very characteristic C<span class="texhtml">α</span> and C<span class="texhtml">β</span> chemical shifts (e.g. Ala, Gly, Ser, Thr) while others can be easily mixed up (Phe/Tyr/Asp/Asn/Cys or Gln/Glu/Met). However, once a few spin systems form a short chain, one can start looking for a proper match with the protein sequence. Again, most software package would suggest best fits.<br> | Some amino acids have very characteristic C<span class="texhtml">α</span> and C<span class="texhtml">β</span> chemical shifts (e.g. Ala, Gly, Ser, Thr) while others can be easily mixed up (Phe/Tyr/Asp/Asn/Cys or Gln/Glu/Met). However, once a few spin systems form a short chain, one can start looking for a proper match with the protein sequence. Again, most software package would suggest best fits.<br> | ||
After the initial match, spin | After the initial match, spin systems linking and sequential backbone assignment often go hand in hand. Once this step is concluded, normally one includes the rest of the side chain nuclei through HCCH and NOESY-type spectra, after which one adds aromatic rings, side chain NH2 groups of Asn and Gln, methyl groups of Met, etc. | ||
Unlike the above process, [[Abacus|ABACUS]] software first expects individual spin systems be as complete as possible, with side chain information included, and | Unlike the above process, [[Abacus|ABACUS]] software first expects individual spin systems be as complete as possible, with side chain information included, and then uses NOESY-type spectra to obtain the assignment via Monte Carlo simulations. | ||
Latest revision as of 21:21, 18 December 2009
Introduction
This section is intended to introduce a few definitions and concepts in resonance assignment protocols. For in depth description of the process see e.g. Kurt Wütrich's book NMR of Proteins and Nucleic Acids (Wiley, 1986) and John Cavangh's et al. textbook Protein NMR Spectroscopy. Principles and Practice (2nd edition, Elsevier, 2007).
Stable isotope labeling schemes
Through the NESG consortium, the most prevalent isotope labeling schemes are as follows:
- 100% 15N, 100%13C-labeled (or doubly-labeled) samples are the main category, to which the majority of the information on this site applies. They are used for complete resonance assignments and structure calculation.
- 100% 15N-labeled samples are used for screening with 15N-HSQC. They can also find limited use in collecting RDC-type data.
- 100% 15N, 5-7% 13C-labeled samples are used to obtain stereospecific assignment of Val and Leu side chain methyl groups, usually important for proper packing of hydrophobic core.
- 100% 14N, 100% 12C (or unlabeled) or alternatively natural abundance samples can be used in 50%-50% mixtures for homodimer structure determination.
In addition to these labeling schemes, one can find it is useful, especially for larger proteins to have selectively labeled samples, such as SAIL NMR (http://www.sailnmr.org/). To reduce signal broadening due to spin-spin relaxation, it may be advantageous to deuterate the protein to a certain level.
NMR experiments
This section describes the types of connectivities that can be established between nuclei by given experiments. For specifics and experimental setup, please refer to the NMR Data Collection section of this site.
Through bond
Through-bond experiments correlate nuclei connected by a limited number of chemical bonds.
- 15N-HSQC - correlates a 15N nucleus and a 1H directly attached to it. Mainly used to identify backbone amide groups.
- 13C-HSQC - correlates a 13C nucleus and a 1H directly attached to it. Can cover aliphatic and/or aromatic range of 13C chemical shift. Constant time version enhances resolution. Peak sign gives information on the number of 13C neighbors a given 13C nucleus has.
- HNCO, 3D - correlates a backbone amide with the C' of preceding residue or a Cγ/Cδ of Asn/Gln with NH2 of the side chain.
- HNCA, 3D - correlates a backbone amide with the Cα of the same and preceding residues.
- HNCACB, 3D - correlates a backbone amide with the Cα and Cβ of the same and preceding residues. Usually has Cβ and Cα peaks of opposite signs.
- CBCA(CO)NH, 3D - correlates a backbone amide with the Cα and Cβ of the preceding residue. Together with HNCACB, HNCA and HNCO, it is used to assign the backbone chemical shifts.
- HBHA(CBCACO)NH, 3D - correlates a backbone amide with the Hα and Hβ resonances of the preceding residue. Used together with HCCH-type experiments and 13C-HSQC to assign the side chain resonances.
- H(C)CH-COSY, 3D - correlates an aliphatic C-H pair with an adjacent aliphatic 1H of the same side chain. Closely related experiment is H(C)CH-TOCSY, 3D, which correlates an aliphatic C-H pair with all aliphatic 1H nuclei of the same side chain.
- (H)CCH-COSY, 3D, and (H)CCH-TOCSY, 3D, are very similar to above, except the third correlated nucleus is 13C.
- All HCCH-type spectra can also be tailored for the aromatic region of 13C range. Correlations would be detected only within an individual aromatic ring.
- (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE, 2D - correlate Hδ and Hε aromatic protons with Cβ of the same residue. Useful for assigning chemical shifts of nuclei in aromatic rings.
- Long range 15N-HSQC, 2D - correlates Hδ2 and Hε1 protons (attached to 12C or 13C nuclei!) on His rings with Nδ1 and Nε2 nuclei. This experiment s useful for assigning Hε1 and Cε1 nuclei of His rings as well as to determine the protonation state of the ring.
Through space
Through-space experiments, as the name implies, correlate pairs of nuclei close in space. Many such nuclei would belong to the same or adjacent residues and these experiments can thus supplement the information obtained from through-bond spectra. The usual distance limit to observe such a correlation is ~5Â.
- 15N-edited NOESY-HSQC, 3D - correlates all protons close to the 1H of an amide group with both nuclei of the amide.
- 13C-edited NOESY-HSQC, 3D - correlates all protons close to the 1H of a C-H moiety with both nuclei of the moiety. Can replace an H(C)CH-TOCSY experiment in certain cases.
The above spectrum can be separated into aliphatic and aromatic regions of the 13C range.
- Simultaneous 13C,15N-edited NOESY-HSQC, 3D - as the name implies, this spectrum combines the above two experiments (15N and 13C).
These NOESY-type spectra are very useful in confirming the sequence-specific backbone assignments and in connecting aromatic rings to their proper side chains. For certain automatic methods (e.g. ABACUS) they are used to establish sequential connectivities so that they can completely replace HNCACB or similar spectra.
Spin systems
Definition
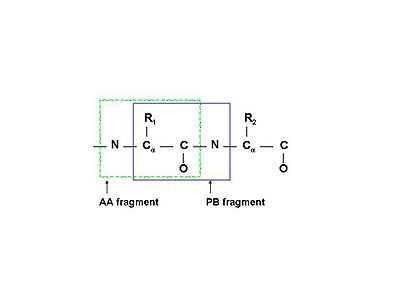
A spin system in the broadest sense of the word is a set of nuclei connected by chemical bonds. While for small molecules this is a very useful definition, for proteins it becomes unwieldy as the whole protein would thus be just one spin system. In practice, in protein NMR, a spin system is usually defined as an amino acid residue (AA-fragment) - N, HN, Cα, Hα, C', and all nuclei of the side chain.
Sometimes it may be more convenient to define a peptide-bond fragment (PB-fragment) spin system, which would contain N and HN of the following amino acid, instead of the same. See figure (left).
Occasionally, an aromatic ring may be treated as a separate spin system from the rest of the same side chain.
Identification
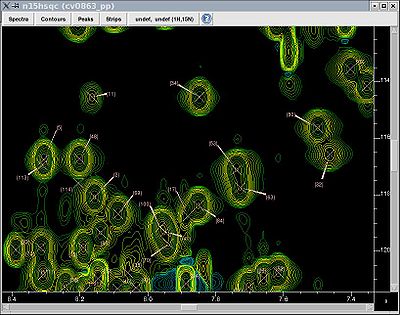
Whether one uses AA- or PB-fragments (see above), a convenient handle on the spin system is the backbone amide group, as there is only one per each residue. Prolines would have no such handle and are treated separately.
One thus normally starts the resonance assignment project with picking the 15N-HSQC spectrum and assigning each peak a unique identifier (Figure, right), which is later used as an identifier of the whole spin system.
Linking spin systems
Once identified, the spin systems can then be linked using information from relevant pairs of spectra, such as HNCACB and CBCA(CO)NH, HN(CO)CA and HNCA, HNCO and HN(CA)CO. Each software package has a slightly different method to do that, but generally for manual linking one looks for identical 13C chemical shifts in intraresidual and sequential spectra, simultaenously displaying strips from the above pairs. The programs often suggest the best fit. Some programs have built-in optimization functions.
Many automatic programs (MONTE, MARS, PINE, AutoAssign) only require peak lists from the above spectra (and 15N-HSQC) and use sofisticated algorithms to link the spin systems sequentially and match them onto the protein sequence.
Matching onto covalent structure
Some amino acids have very characteristic Cα and Cβ chemical shifts (e.g. Ala, Gly, Ser, Thr) while others can be easily mixed up (Phe/Tyr/Asp/Asn/Cys or Gln/Glu/Met). However, once a few spin systems form a short chain, one can start looking for a proper match with the protein sequence. Again, most software package would suggest best fits.
After the initial match, spin systems linking and sequential backbone assignment often go hand in hand. Once this step is concluded, normally one includes the rest of the side chain nuclei through HCCH and NOESY-type spectra, after which one adds aromatic rings, side chain NH2 groups of Asn and Gln, methyl groups of Met, etc.
Unlike the above process, ABACUS software first expects individual spin systems be as complete as possible, with side chain information included, and then uses NOESY-type spectra to obtain the assignment via Monte Carlo simulations.