Processing non-uniformly sampled spectra with Multidimensional Decomposition: Difference between revisions
(Created page with '=== Processing with MDD === This section gives a step-by-step manual to processing with the MDDGUI program, developed in the Arrowsmith lab (University Health Network and Unive…') |
|||
| (72 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== | __NORICHEDITOR__ | ||
== Introduction and Prerequisites == | |||
This section gives a step-by-step manual to processing with the MDDGUI program, developed in the Arrowsmith lab (University Health Network and University of Toronto). It is designed for processing 3D spectra collected with non-uniform or non-linear sampling (NUS). The spectra can be collected either on Varian spectrometers with later versions of BioPack installed ([[NUS guide for Varian|click here for the relevant guide for Varian spectrometers]]) or on Bruker spectrometers running TopSpin ([[NUS guide for Bruker according to Arrowsmith group in Toronto|click here for the Arrowsmith protocol for NUS data collection on Bruker instruments]]). | This section gives a step-by-step manual to processing with the MDDGUI program, developed in the Arrowsmith lab (University Health Network and University of Toronto). It is designed for processing 3D spectra collected with non-uniform or non-linear sampling (NUS). The spectra can be collected either on Varian spectrometers with later versions of BioPack installed ([[Setting up non-uniformly sampled spectra/NUS guide for Varian|click here for the relevant guide for Varian spectrometers]]) or on Bruker spectrometers running TopSpin ([[Setting up non-uniformly sampled spectra/NUS guide for Bruker according to Arrowsmith group in Toronto|click here for the Arrowsmith protocol for NUS data collection on Bruker instruments]]). | ||
The GUI package | The GUI package requires the '''mddnmr''' software developed by [http://www.nmr.gu.se/english/research/orekhov Vladislav Orekhov] (Swedish NMR Centre, University of Gothenburg). Please contact him for your copy of the software. The GUI can be requested from the Arrowsmith group. You need Qt libraries in order to compile the GUI, or you can use the precompiled Linux version. Please refer to the installation README file for details. In addition to these two packages, you should have working nmrPipe and nmrDraw programs, as apodization, Fourier transforms, etc are performed by nmrPipe. | ||
__NORICHEDITOR__ | |||
== Processing with MDDGUI == | |||
__NORICHEDITOR__ | |||
Before the Multi-dimensional decomposition can be started, the raw data has to be converted through a number of steps to an appropriate format. The steps below are accompanied by the GUI screen shots. To move between the different stages of the processing, use "Next" and "Back" buttons. Refer to README file or tutorial for more specific instructions. | |||
<br> | |||
__NORICHEDITOR__ | |||
=== Step 1. Reshuffle FIDs === | |||
__NORICHEDITOR__ | |||
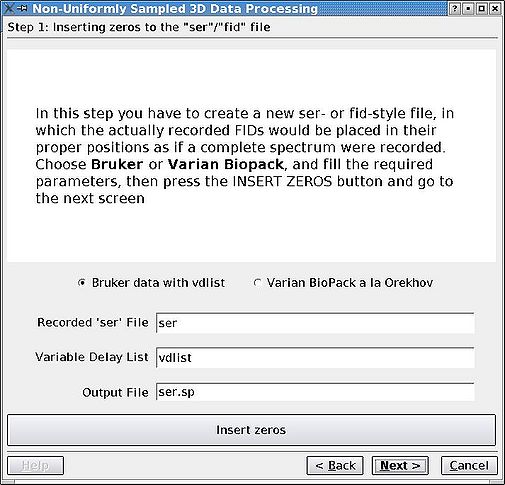
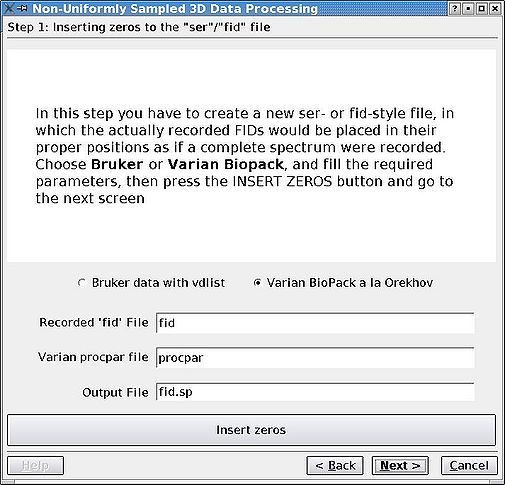
Rearrange the FIDs in the '''ser''' of '''fid''' file so that they are in correct positions for an imaginary case as if the data set was sampled regularly. The missing FIDs are replaced by zeros as dictated by the vdlist or procpar files. First, click the appropriate radio button, edit any file names if necessary, and click '''Insert Zeros''' before proceeding. <br>[[Image:MDDGUIstep1.jpg|thumb|left|505px|Figure 1a. Reshuffling FIDs in Bruker <tt>ser</tt> file.]][[Image:MDDGUIstep1 Varian.jpg|thumb|right|505px|Figure 1b. Reshuffling FIDs in Varian <tt>fid</tt> file.]] | |||
<br style="clear: both" /> | |||
<br> <br> | |||
__NORICHEDITOR__ | |||
=== Step 2. Convert to NMRPipe format === | |||
__NORICHEDITOR__ | |||
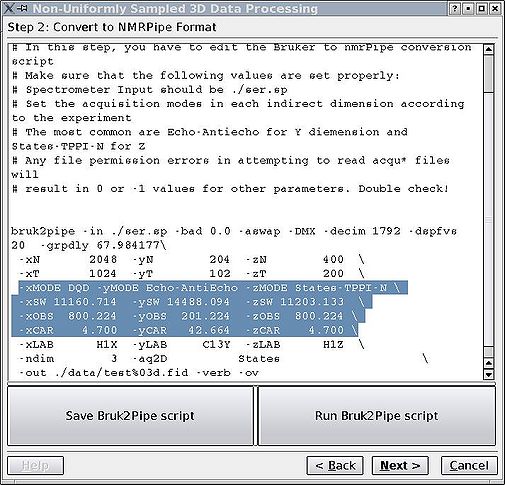
In this step there are a few parameters that the GUI might read wrong, especially with later versions of TopSpin. You can edit it as a simple bruk2pipe or var2pipe script. Pay extra attention to carrier and spectrometer frequencies, matrix sizes and acquisition modes (highlighted). As far as indirect dimensions' acquisition modes are concerned, it is important to distinguish between Echo-AntiEcho (Rance-Kay) and Complex/States-TPPI modes, as one can select within the group by adjusting flags for "nmrPipe -fn FT" command. In TopSpin2.1 we discovered that sweep width is sometimes not recorded in the acqu* files, therefore check that as well. Click '''Save''', then '''Run'''. <br> | |||
<br> | |||
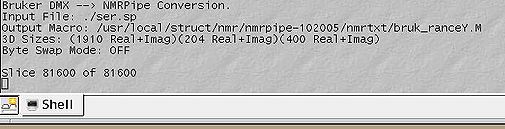
<br>[[Image:MDDGUIstep2.jpg|thumb|left|505px|Figure 2a. Making sure the highlighted parameters are correct]][[Image:MDDGUIstep2 out.jpg|thumb|right|505px|Figure 2b. Output of the conversion to nmrPipe format]] NB! Make sure the spectrometer input is the reshuffled file (ser.sp or fid.sp)! <br> <br> | |||
<br style="clear: both" /> | |||
__NORICHEDITOR__ | |||
=== Step 3. Process the first plane <br> === | |||
__NORICHEDITOR__ | |||
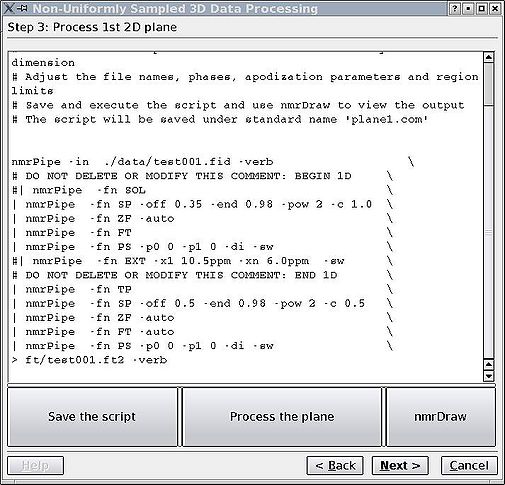
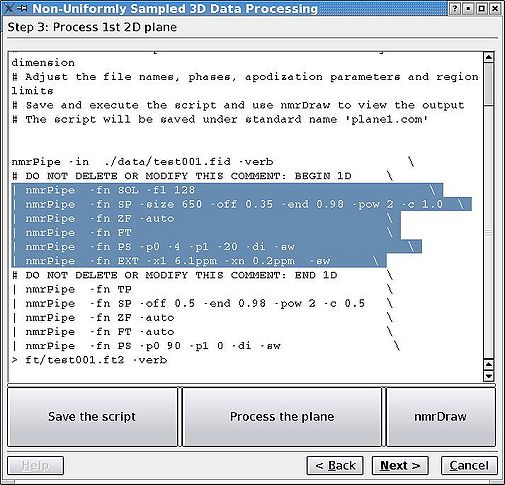
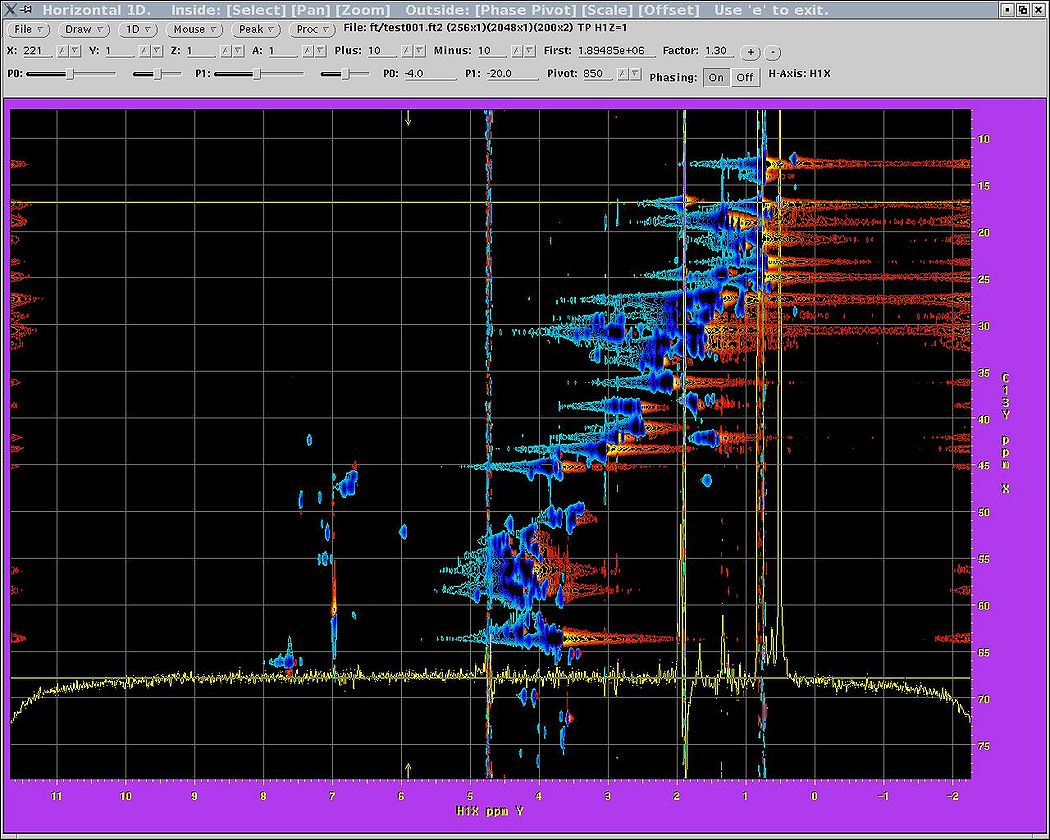
& | For the Bruker data, the first plane is regularly sampled while for Varian it is not, which may make this step more difficult for Varian data. One only needs to adjust phasing, apodisation, region of interest and other parameters for the '''acquisition '''or directly detected dimension. The better the phasing, the better the convergence and the fewer the artefacts. It will be difficult to change the acquisition dimension parameters afterwards without redoing the calculation, so spend a few rounds with NMRDraw until you are happy and the phases are set properly (see highlighted section in the figure). As usual, click "Save", then "Run" each time you want to reprocess the plane. Button "nmrDraw" simply calls that program. | ||
<br> [[Image:MDDGUIstep3.jpg|thumb|left|505px|Figure 3a. Initial script to process the first plane of the spectrum.]][[Image:MDDGUIstep3 final.jpg|thumb|right|505px|Figure 3b. After the adjustment of relevant parameters.]] [[Image:MDDGUIstep3 plane.jpg|thumb|center|1050px|Figure 3c. NMRDraw screenshot for phasing the acquisition dimension.]] <br> | |||
<br style="clear: both" /> | |||
__NORICHEDITOR__ | |||
=== Step 4. Processing the acquisition dimension<br> === | |||
__NORICHEDITOR__ | |||
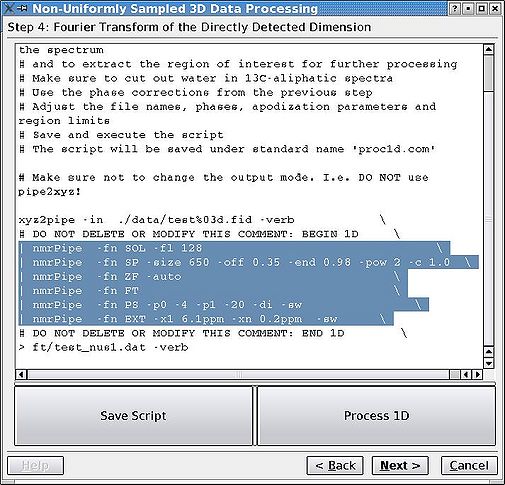
Using the parameters from step 3, Fourier transform (FT) the acquisition dimension only for the whole data set and extract the region of interest. Note that the parameters are copied, but one can still edit them before proceeding. <br> [[Image:MDDGUIstep4.jpg|thumb|center|505px| Figure 4. Processing the acquisition dimension.]]<br> | |||
<br style="clear: both" /> | |||
__NORICHEDITOR__ | |||
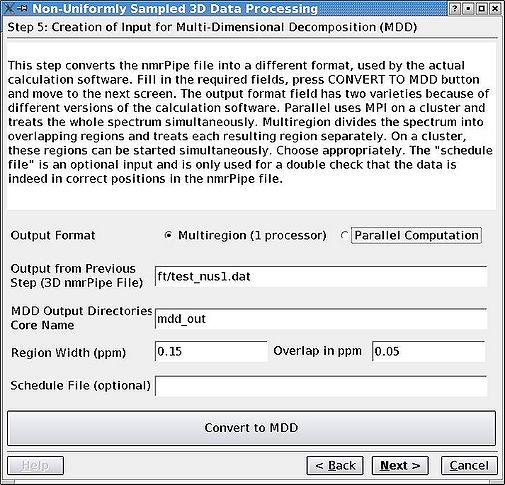
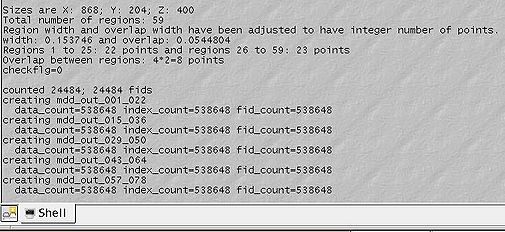
=== Step 5. Converting to MDDNMR format<br> === | |||
__NORICHEDITOR__ | |||
Divide the the partially Fourier transformeded but still sparse spectrum into a number of regions overlapping by their acquisition dimension and convert these regions to MDDNMR format. This is the step before proceeding to calculations themselves. Make sure you select "Multiregion" radiobutton. Parallel computation is becoming obsolete, although is supported for now. Region width should be given in ppm | |||
<br> [[Image:MDDGUIstep5.jpg|thumb|left|505px|Figure 5a. Slicing the spectrum into overlapping regions and converting to MDDNMR format.]][[Image:MDDGUIstep5 out.jpg|thumb|right|505px|Figure 5b. Output of the conversion to MDDNMR format.]] <br> <br> | |||
<br style="clear: both" /> | |||
__NORICHEDITOR__ | |||
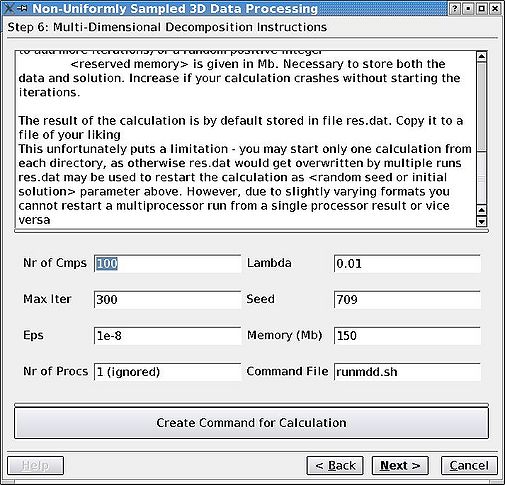
=== Step 6. Run MDD <br> === | |||
__NORICHEDITOR__ | |||
As simple as that. Check README for more detailed instructions. The most important parameter is the number of components (per region), which should be the maximum number of HSQC/projection peaks . You might need to do some editing of the "runmdd.sh" script depending on cluster availability (''e.g.'', you may need to remove & from the mddnmr command if your calculation is running on a desktop). In the Arrowsmith group, we usually open a separate terminal window, login to our cluster and start the calculations from there. '''Do not close the GUI''' in the meantime. If you do, it is a bit tricky to restart from where you are. | |||
<br> [[Image:MDDGUIstep6.jpg|thumb|left|505px|Figure 6a. Parameters for the MDDNMR run.]] [[Image:MDDGUIstep6 run.jpg|thumb|right|505px|Figure 6b. How to start the calculation on a cluster.]] | |||
<br style="clear: both" /> | |||
__NORICHEDITOR__ | |||
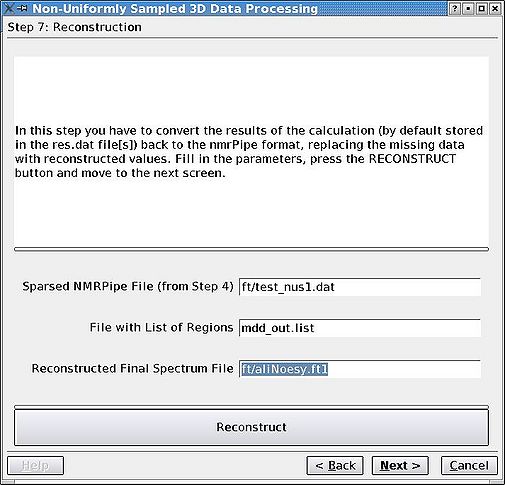
=== Step 7. Reconstructing the spectrum === | |||
__NORICHEDITOR__ | |||
Using the output of MDD, replace the zeros ( ''i.e.'' missing FIDs) by the reconstructed values. Here, if you had the GUI open through the previous step, the values will be filled automatically. If you did close it, restart it, go to step 5, choose "Multiregion" radiobutton, proceed to step 7 and fill out the values. That was the tricky part. | |||
<br> [[Image:MDDGUIstep7.jpg|thumb|left|505px|Figure 7a. Reconstructing the full spectrum.]] [[Image:MDDGUIstep7 out.jpg|thumb|right|505px|Figure 7b. Output messages from the reconstruction step.]] | |||
<br> | |||
<br style="clear: both" /> | |||
__NORICHEDITOR__ | |||
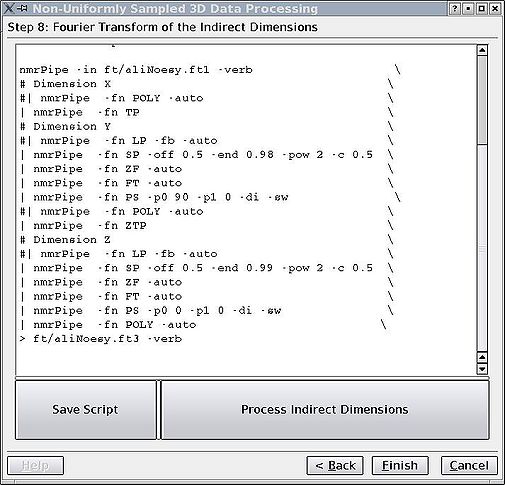
=== Step 8. Fourier transform of indirect dimensions === | |||
__NORICHEDITOR__ | |||
This is the final step of the processing. Here the task is purely NMRPipe. You may do to your indirect dimensions what you need to do - apodization, baseline correction, etc, etc. You may need to play around with "FT -auto" if your peaks are not making sense. That is mainly because the GUI can't easily distinguish between "Complex", "States-TPPI" and related modes. So you may need to change the FT flags to rectify the situation. | |||
{| border="1" | |||
|+ '''Correspondence of FT flags to Complex acquisition modes''' | |||
|- | |||
! FT flag(s) | |||
! Aquisition mode | |||
! Action(s) performed | |||
! Effect(s) on spectrum | |||
|- | |||
| FT | |||
| Complex | |||
| none | |||
| none | |||
|- | |||
| FT -alt | |||
| States-TPPI | |||
| sign alternation for every other point | |||
| Left and right (top and bottom) halves swapped | |||
|- | |||
| FT -neg | |||
| Complex-N | |||
| negation of imaginaries | |||
| spectrum inverted | |||
|- | |||
| FT -alt -neg | |||
| States-TPPI-N | |||
| sign alternation and negation of imaginaries | |||
| spectrum halves swapped and the whole spectrum inverted | |||
|} | |||
Finally, you don't need the GUI anymore, as you can edit the script to your liking with any text editor. | |||
<br> [[Image:MDDGUIstep8.jpg|thumb|center|505px|Figure 8. Finalizing the Fourier transform of the indirect dimensions.]] | |||
<br> | |||
<br style="clear: both" /> | |||
__NORICHEDITOR__ | |||
== Cleaning up == | |||
Once you have processed your spectrum, there are a lot of temporary files that are no longer needed. By deleting them you can sometimes free up to a Gb of disk space. If you leave the standard names for these files you can delete the following: | |||
{| | |||
|- | |||
! Bruker | |||
! Varian | |||
|- | |||
| <pre> > rm -rf data/ pdata/ mdd_out_* ser.sp ft/test_nus1.dat | |||
> gzip ser</pre> | |||
| <pre> > rm -rf data/ mdd_out_* fid.sp ft/test_nus1.dat | |||
> gzip fid </pre> | |||
|} | |||
You should, of course, modify accordingly, if your file names are different. In this particular example, one could also delete the ft/aliNoesy.ft1 file. Note also, that if you perform the processing on the spectrometer, it is desirable to keep the pdata/ directory intact. | |||
<br> | |||
== Software requests == | |||
<br> | |||
*MDDGUI package can be requested from the Arrowsmith group or by e-mailing (gutmanas@ebi.ac.uk) | |||
*Mddnmr software is developed by [http://www.nmr.gu.se/english/research/orekhov Vladislav Orekhov] (Swedish NMR Centre, University of Gothenburg). Please contact him for your copy of the package. <br> | |||
Latest revision as of 12:54, 26 July 2010
__NORICHEDITOR__
Introduction and Prerequisites
This section gives a step-by-step manual to processing with the MDDGUI program, developed in the Arrowsmith lab (University Health Network and University of Toronto). It is designed for processing 3D spectra collected with non-uniform or non-linear sampling (NUS). The spectra can be collected either on Varian spectrometers with later versions of BioPack installed (click here for the relevant guide for Varian spectrometers) or on Bruker spectrometers running TopSpin (click here for the Arrowsmith protocol for NUS data collection on Bruker instruments).
The GUI package requires the mddnmr software developed by Vladislav Orekhov (Swedish NMR Centre, University of Gothenburg). Please contact him for your copy of the software. The GUI can be requested from the Arrowsmith group. You need Qt libraries in order to compile the GUI, or you can use the precompiled Linux version. Please refer to the installation README file for details. In addition to these two packages, you should have working nmrPipe and nmrDraw programs, as apodization, Fourier transforms, etc are performed by nmrPipe.
__NORICHEDITOR__
Processing with MDDGUI
__NORICHEDITOR__
Before the Multi-dimensional decomposition can be started, the raw data has to be converted through a number of steps to an appropriate format. The steps below are accompanied by the GUI screen shots. To move between the different stages of the processing, use "Next" and "Back" buttons. Refer to README file or tutorial for more specific instructions.
__NORICHEDITOR__
Step 1. Reshuffle FIDs
__NORICHEDITOR__
Rearrange the FIDs in the ser of fid file so that they are in correct positions for an imaginary case as if the data set was sampled regularly. The missing FIDs are replaced by zeros as dictated by the vdlist or procpar files. First, click the appropriate radio button, edit any file names if necessary, and click Insert Zeros before proceeding.
__NORICHEDITOR__
Step 2. Convert to NMRPipe format
__NORICHEDITOR__
In this step there are a few parameters that the GUI might read wrong, especially with later versions of TopSpin. You can edit it as a simple bruk2pipe or var2pipe script. Pay extra attention to carrier and spectrometer frequencies, matrix sizes and acquisition modes (highlighted). As far as indirect dimensions' acquisition modes are concerned, it is important to distinguish between Echo-AntiEcho (Rance-Kay) and Complex/States-TPPI modes, as one can select within the group by adjusting flags for "nmrPipe -fn FT" command. In TopSpin2.1 we discovered that sweep width is sometimes not recorded in the acqu* files, therefore check that as well. Click Save, then Run.
NB! Make sure the spectrometer input is the reshuffled file (ser.sp or fid.sp)!
__NORICHEDITOR__
Step 3. Process the first plane
__NORICHEDITOR__
For the Bruker data, the first plane is regularly sampled while for Varian it is not, which may make this step more difficult for Varian data. One only needs to adjust phasing, apodisation, region of interest and other parameters for the acquisition or directly detected dimension. The better the phasing, the better the convergence and the fewer the artefacts. It will be difficult to change the acquisition dimension parameters afterwards without redoing the calculation, so spend a few rounds with NMRDraw until you are happy and the phases are set properly (see highlighted section in the figure). As usual, click "Save", then "Run" each time you want to reprocess the plane. Button "nmrDraw" simply calls that program.
__NORICHEDITOR__
Step 4. Processing the acquisition dimension
__NORICHEDITOR__
Using the parameters from step 3, Fourier transform (FT) the acquisition dimension only for the whole data set and extract the region of interest. Note that the parameters are copied, but one can still edit them before proceeding.
__NORICHEDITOR__
Step 5. Converting to MDDNMR format
__NORICHEDITOR__
Divide the the partially Fourier transformeded but still sparse spectrum into a number of regions overlapping by their acquisition dimension and convert these regions to MDDNMR format. This is the step before proceeding to calculations themselves. Make sure you select "Multiregion" radiobutton. Parallel computation is becoming obsolete, although is supported for now. Region width should be given in ppm
__NORICHEDITOR__
Step 6. Run MDD
__NORICHEDITOR__
As simple as that. Check README for more detailed instructions. The most important parameter is the number of components (per region), which should be the maximum number of HSQC/projection peaks . You might need to do some editing of the "runmdd.sh" script depending on cluster availability (e.g., you may need to remove & from the mddnmr command if your calculation is running on a desktop). In the Arrowsmith group, we usually open a separate terminal window, login to our cluster and start the calculations from there. Do not close the GUI in the meantime. If you do, it is a bit tricky to restart from where you are.
__NORICHEDITOR__
Step 7. Reconstructing the spectrum
__NORICHEDITOR__
Using the output of MDD, replace the zeros ( i.e. missing FIDs) by the reconstructed values. Here, if you had the GUI open through the previous step, the values will be filled automatically. If you did close it, restart it, go to step 5, choose "Multiregion" radiobutton, proceed to step 7 and fill out the values. That was the tricky part.
__NORICHEDITOR__
Step 8. Fourier transform of indirect dimensions
__NORICHEDITOR__
This is the final step of the processing. Here the task is purely NMRPipe. You may do to your indirect dimensions what you need to do - apodization, baseline correction, etc, etc. You may need to play around with "FT -auto" if your peaks are not making sense. That is mainly because the GUI can't easily distinguish between "Complex", "States-TPPI" and related modes. So you may need to change the FT flags to rectify the situation.
| FT flag(s) | Aquisition mode | Action(s) performed | Effect(s) on spectrum |
|---|---|---|---|
| FT | Complex | none | none |
| FT -alt | States-TPPI | sign alternation for every other point | Left and right (top and bottom) halves swapped |
| FT -neg | Complex-N | negation of imaginaries | spectrum inverted |
| FT -alt -neg | States-TPPI-N | sign alternation and negation of imaginaries | spectrum halves swapped and the whole spectrum inverted |
Finally, you don't need the GUI anymore, as you can edit the script to your liking with any text editor.
__NORICHEDITOR__
Cleaning up
Once you have processed your spectrum, there are a lot of temporary files that are no longer needed. By deleting them you can sometimes free up to a Gb of disk space. If you leave the standard names for these files you can delete the following:
| Bruker | Varian |
|---|---|
> rm -rf data/ pdata/ mdd_out_* ser.sp ft/test_nus1.dat > gzip ser |
> rm -rf data/ mdd_out_* fid.sp ft/test_nus1.dat > gzip fid |
You should, of course, modify accordingly, if your file names are different. In this particular example, one could also delete the ft/aliNoesy.ft1 file. Note also, that if you perform the processing on the spectrometer, it is desirable to keep the pdata/ directory intact.
Software requests
- MDDGUI package can be requested from the Arrowsmith group or by e-mailing (gutmanas@ebi.ac.uk)
- Mddnmr software is developed by Vladislav Orekhov (Swedish NMR Centre, University of Gothenburg). Please contact him for your copy of the package.