Setting up non-uniformly sampled spectra/NUS guide for Bruker according to Arrowsmith group in Toronto: Difference between revisions
No edit summary |
|||
| (27 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__NORICHEDITOR__ | |||
__TOC__ | |||
== Setting up acquisition of non-uniformly sampled 3D spectra on Bruker spectrometers with TopSpin == | |||
__NORICHEDITOR__ | |||
A standardized way to set non-uniform sampling (NUS) for TopSpin is in the making, however, while it is not yet included in the current distribution, this page will focus on the way implemented by the Arrowsmith group in Toronto (University | A standardized way to set non-uniform sampling (NUS) for TopSpin is in the making, however, while it is not yet included in the current distribution, this page will focus on the way Non-uniform sampling of 3D spectra is implemented by the Arrowsmith group in Toronto (University Health Network and University of Toronto). It is tailored for the subsequent processing with MDDGUI software, described elsewhere on this site.<br> | ||
To set up 3D NUS experiments according to this method one needs the following:<br> | To set up 3D NUS experiments according to this method one needs the following:<br> | ||
| Line 9: | Line 12: | ||
*A Jython script for generating the sampling schedule<br> | *A Jython script for generating the sampling schedule<br> | ||
__NORICHEDITOR__ | |||
=== Pulse program modifications<br> === | |||
__NORICHEDITOR__ | |||
First of all, one has to set up a regular experiment with all pulses and delays calibrated as usual. Once this step is finished, one needs to replace the pulse program by a modified one. | First of all, one has to set up a regular experiment with all pulses and delays calibrated as usual. Once this step is finished, one needs to replace the pulse program by a modified one. | ||
This section briefly describes the essential pulse sequence modifications. The | This section briefly describes the essential pulse sequence modifications. The <sup>15</sup>N-NOESY-HSQC pulse program is used as an example. In most cases one just need to copy these changes into relevant pulse program.<br> | ||
*Additional definitions should come with other definitions in the file:<br> | *Additional definitions should come with other definitions in the file:<br> | ||
| Line 23: | Line 28: | ||
;^^^^^^^^^^^^^^^^^^^^^^^^^^^^ | ;^^^^^^^^^^^^^^^^^^^^^^^^^^^^ | ||
</pre> | </pre> | ||
*Main '''if''' statement determines whether an Echo/AntiEcho or Real/Imaginary pair is recorded or skipped. The information is taken from the '''vdlist''' file | *Main '''if''' statement determines whether an Echo/AntiEcho or Real/Imaginary pair is recorded or skipped. The information is taken from the '''vdlist''' file which is generated by a separate script before running the experiments. If the current value of '''vd''' is 1u, the pair is skipped (goto 100 statement), if it is 3u, it is recorded. This addition replaces the text commented out and comes before the '''d1''' delay:<br> | ||
<pre>;1 ze | <pre>;1 ze | ||
; d11 pl16:f3 | ; d11 pl16:f3 | ||
| Line 75: | Line 80: | ||
exit | exit | ||
</pre> | </pre> | ||
A good number of pulse programs are already modified and are available | A good number of pulse programs are already modified and are available upon request (see [[Setting up non-uniformly sampled spectra]]): | ||
{| cellspacing="1" | |||
| | |||
* HNCO (hncogp3d.nus) | |||
* HNCA (hncagp3d.nus) | |||
* CBCA(CO)NH (cbcaconhgp3d.nus) | |||
* HBHA(CO)NH (hbhaconhgp3d.nus) | |||
| | |||
* <sup>15</sup>N-NOESY-HSQC (noesyhsqcfpf3gpsi3d.nus) | |||
* <sup>13</sup>C-NOESY-HSQC (noesyhsqcetgp3d.nus) | |||
* H(C)CH-TOCSY (hcchdigp3d3.nus) | |||
* (H)CCH-TOCSY (hcchdigp3d2.nus) | |||
|} | |||
Once the experiment is otherwise set up and the pulse program modifications are in place, the last step before running the experiment is to generate the sampling schedule. Depending on whether your computer runs Linux or Windows, the generator differs slightly (mainly in the usage of forward and backslashes). Both scripts are available | __NORICHEDITOR__ | ||
=== Generating the sampling schedule and running the experiment === | |||
__NORICHEDITOR__ | |||
Once the experiment is otherwise set up and the pulse program modifications are in place, the last step before running the experiment is to generate the sampling schedule. Depending on whether your computer runs Linux or Windows, the generator differs slightly (mainly in the usage of forward and backslashes). Both scripts are available upon request from the Arrowsmith group (together with the above pulse programs). The corresponding script should be renamed '''sparse.py''' and put in '''/opt/topspin/exp/stan/nmr/py/user/''' (Linux) or '''C:\Bruker\TOPSPIN\exp\stan\nmr\py\user\''' (Windows) on your computer (NB! The directory may be slightly different on your system).<br> | |||
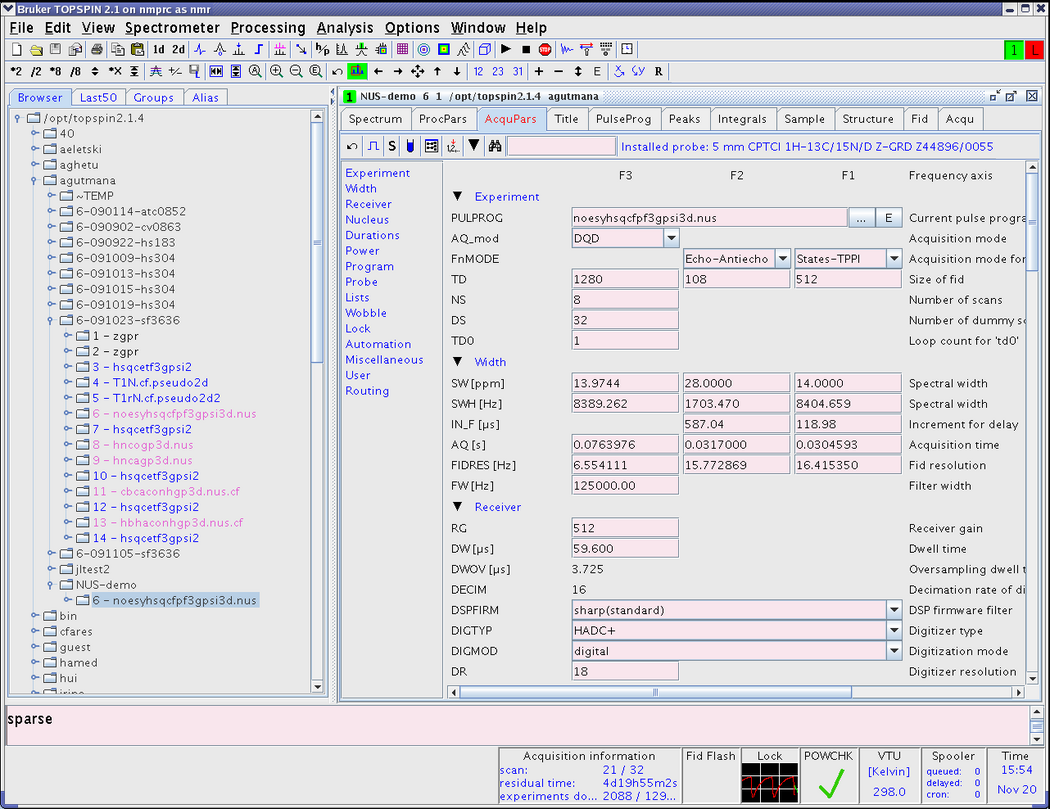
It can be called by typing '''sparse''' from the command prompt. | It can be called by typing '''sparse''' from the command prompt. | ||
[[Image:TopspinNUSSnap0.png|thumb|center|1050px]] | [[Image:TopspinNUSSnap0.png|thumb|center|1050px|Figure 1. Changing the pulse program and calling sparse.py script]] | ||
After the initial greeting, you will be guided through a few steps as shown below. | After the initial greeting, you will be guided through a few steps as shown below. | ||
| Line 89: | Line 110: | ||
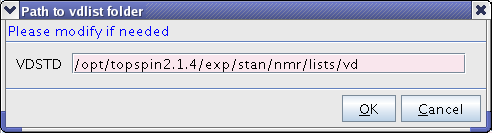
*All vdlist files should be copied to a standard directory. If the suggested directory is wrong, please modify. | *All vdlist files should be copied to a standard directory. If the suggested directory is wrong, please modify. | ||
[[Image:TopspinNUSSnap2.png|center]] | [[Image:TopspinNUSSnap2.png|frame|center|400px|Figure 2. Where the new vdlist should be placed.]] | ||
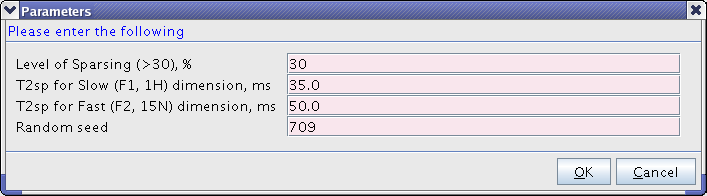
*Main relevant parameters are presented in this window.The level of "sparsing" allows to fine tune the acquisition time. It should generally be 30% or more. T2sp parameters dictate the exponential biasing of the sampling and only need to be approximate. In the Arrowsmith group we use 20ms for 13C, 35 ms for 1H and 50ms for 15N. Usually the values are guessed correctly, but you can modify them if desired. | *Main relevant parameters are presented in this window. The level of "sparsing" allows to fine tune the acquisition time. It should generally be 30% or more. T2sp parameters dictate the exponential biasing of the sampling and only need to be approximate. In the Arrowsmith group we use 20ms for 13C, 35 ms for 1H and 50ms for 15N. Usually the values are guessed correctly, but you can modify them if desired. | ||
[[Image:TopspinNUSSnap3.png|center]] | [[Image:TopspinNUSSnap3.png|frame|center|620px|Figure 3. Providing the parameters. Level of "sparsing" will give the number of FIDs actually recorded.]] | ||
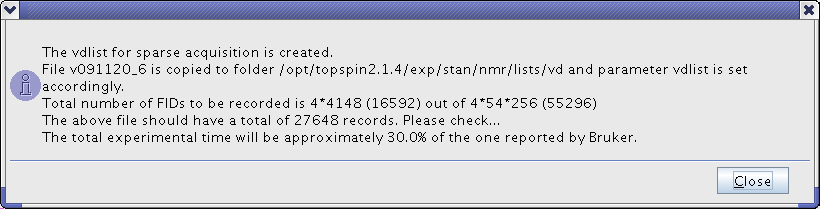
*Concluding window shows the final information on the number of FIDs to be recorded. Note that the experimental time shown by expt command will be wrong and should be multiplied by the "level of sparsing" parameter to give a correct estimate. | *Concluding window shows the final information on the number of FIDs to be recorded. Note that the experimental time shown by expt command will be wrong and should be multiplied by the "level of sparsing" parameter to give a correct estimate. | ||
[[Image:TopspinNUSSnap4.png|center]] | [[Image:TopspinNUSSnap4.png|frame|center|Figure 4. Checking that the file is copied to the right place and that the number of FIDs makes sense.]] | ||
*To run the experiment type '''zg''' or '''multizg''' as the case may be. To process the spectrum you will need the '''vdlist''' file in addition to the usual parameter files. | |||
=== Special Remarks === | |||
The script and the sampling schedule have a few features worthy of some remarks: | |||
*The first increments in each indirect dimension are sampled completely. This assists later in processing the data. | |||
*The first plane in the "fast" dimension (usually 23 or XY) can be processed with TOPSPIN as if you were running a regular spectrum (i.e. with xfb). | |||
*The first plane of the "slow" dimension (usually 13 or XZ) however cannot be processed in TOPSPIN unless the raw data is unscrambled first. | |||
*Command expt gives incorrect experimental time estimate (see the final step above). | |||
*An interrupted experiment cannot be continued by typing "go". | |||
*The generated vdlist is named by default as follows "v"+date+"_"+ExpNum. Usually this should not cause problems, unless you wish to run two NUS experiments on the same day in the same experiment folder (number). | |||
*The vdlist should have TD<sub>Y</sub>*TD<sub>Z</sub>/2 entries as each entry corresponds to recording or skipping two FIDs (Re/Im or Echo/AntiEcho). Bruker convention to record all increments in the fast dimension and only then increment the slow means that the selection for each slow plane appears twice (once for Real and once for Imaginary point to ensure proper quadrature detection in the slow indirect dimension). This is the reason you should rerun the script if you change these two parameters, as otherwise the data will be difficult to unscramble.<br> | |||
*The 2 T2sp parameters give the exponential bias of the sampling in the two indirect dimensions. We did not investigate whether this value has an effect on dimensions with constant time evolution. | |||
Latest revision as of 16:56, 3 November 2010
__NORICHEDITOR__
Setting up acquisition of non-uniformly sampled 3D spectra on Bruker spectrometers with TopSpin
__NORICHEDITOR__
A standardized way to set non-uniform sampling (NUS) for TopSpin is in the making, however, while it is not yet included in the current distribution, this page will focus on the way Non-uniform sampling of 3D spectra is implemented by the Arrowsmith group in Toronto (University Health Network and University of Toronto). It is tailored for the subsequent processing with MDDGUI software, described elsewhere on this site.
To set up 3D NUS experiments according to this method one needs the following:
- Modified pulse program(s)
- A Jython script for generating the sampling schedule
__NORICHEDITOR__
Pulse program modifications
__NORICHEDITOR__
First of all, one has to set up a regular experiment with all pulses and delays calibrated as usual. Once this step is finished, one needs to replace the pulse program by a modified one.
This section briefly describes the essential pulse sequence modifications. The 15N-NOESY-HSQC pulse program is used as an example. In most cases one just need to copy these changes into relevant pulse program.
- Additional definitions should come with other definitions in the file:
;********Added for NUS******* define delay short "short=50u" "l2=td2/2" "l3=td1/2" ;^^^^^^^^^^^^^^^^^^^^^^^^^^^^
- Main if statement determines whether an Echo/AntiEcho or Real/Imaginary pair is recorded or skipped. The information is taken from the vdlist file which is generated by a separate script before running the experiments. If the current value of vd is 1u, the pair is skipped (goto 100 statement), if it is 3u, it is recorded. This addition replaces the text commented out and comes before the d1 delay:
;1 ze ; d11 pl16:f3 ;2 d11 do:f3 ;3 d12 ;********Added for NUS******* 1 ze d11 pl16:f3 2 d11 do:f3 short*2 3 short*5 4 1u if "vd < 2u" goto 100 short*6 5 short 6 d11 ;^^^^^^^^^^^^^^^^^^^^^^^^^^^^ d1
- The final modification comes at the end of the pulse program and uses explicit looping (old style Bruker pulse programming). Note that F1PH and F2EA statements are commented out and the phase and delay increments or decrements are stated explicitly. These increments or decrements need to be executed even if the particular fids are skipped
go=2 ph31 cpd3:f3 ;********Added for NUS******* d11 do:f3 wr #0 if #0 zd ;---------------------------- 100 short igrad EA ; even if the pair is skipped, phases have to be increment appropriately. short ip5*2 ; especially important if Complex or States-TPPI mode used for this dimension lo to 3 times 2 ; end of Echo-AntiEcho loop short id10 ; increment delay and phases for 15N short ip3*2 short ip6*2 short ip31*2 short ivd ; check the next value of vd lo to 4 times l2 ; go back to beginng with the new vd value short rd10 ; reset 15N delays and phases short rp3 short rp6 short rp31 short ip8 ; increment 1H-indirect phases short ip9 lo to 5 times 2 short id0 ; increment 1H-indirect delay. lo to 6 times l3 1m ;^^^^^^^^^^^^^^^^^^^^^^^^^^^^ ; d11 do:f3 mc #0 to 2 ; F1PH(rd10 & rp3 & rp6 & rp31 & ip8 & ip9, id0) ; F2EA(igrad EA & ip5*2, id10 & ip3*2 & ip6*2 & ip31*2) exit
A good number of pulse programs are already modified and are available upon request (see Setting up non-uniformly sampled spectra):
|
|
__NORICHEDITOR__
Generating the sampling schedule and running the experiment
__NORICHEDITOR__
Once the experiment is otherwise set up and the pulse program modifications are in place, the last step before running the experiment is to generate the sampling schedule. Depending on whether your computer runs Linux or Windows, the generator differs slightly (mainly in the usage of forward and backslashes). Both scripts are available upon request from the Arrowsmith group (together with the above pulse programs). The corresponding script should be renamed sparse.py and put in /opt/topspin/exp/stan/nmr/py/user/ (Linux) or C:\Bruker\TOPSPIN\exp\stan\nmr\py\user\ (Windows) on your computer (NB! The directory may be slightly different on your system).
It can be called by typing sparse from the command prompt.
After the initial greeting, you will be guided through a few steps as shown below.
- All vdlist files should be copied to a standard directory. If the suggested directory is wrong, please modify.
- Main relevant parameters are presented in this window. The level of "sparsing" allows to fine tune the acquisition time. It should generally be 30% or more. T2sp parameters dictate the exponential biasing of the sampling and only need to be approximate. In the Arrowsmith group we use 20ms for 13C, 35 ms for 1H and 50ms for 15N. Usually the values are guessed correctly, but you can modify them if desired.
- Concluding window shows the final information on the number of FIDs to be recorded. Note that the experimental time shown by expt command will be wrong and should be multiplied by the "level of sparsing" parameter to give a correct estimate.
- To run the experiment type zg or multizg as the case may be. To process the spectrum you will need the vdlist file in addition to the usual parameter files.
Special Remarks
The script and the sampling schedule have a few features worthy of some remarks:
- The first increments in each indirect dimension are sampled completely. This assists later in processing the data.
- The first plane in the "fast" dimension (usually 23 or XY) can be processed with TOPSPIN as if you were running a regular spectrum (i.e. with xfb).
- The first plane of the "slow" dimension (usually 13 or XZ) however cannot be processed in TOPSPIN unless the raw data is unscrambled first.
- Command expt gives incorrect experimental time estimate (see the final step above).
- An interrupted experiment cannot be continued by typing "go".
- The generated vdlist is named by default as follows "v"+date+"_"+ExpNum. Usually this should not cause problems, unless you wish to run two NUS experiments on the same day in the same experiment folder (number).
- The vdlist should have TDY*TDZ/2 entries as each entry corresponds to recording or skipping two FIDs (Re/Im or Echo/AntiEcho). Bruker convention to record all increments in the fast dimension and only then increment the slow means that the selection for each slow plane appears twice (once for Real and once for Imaginary point to ensure proper quadrature detection in the slow indirect dimension). This is the reason you should rerun the script if you change these two parameters, as otherwise the data will be difficult to unscramble.
- The 2 T2sp parameters give the exponential bias of the sampling in the two indirect dimensions. We did not investigate whether this value has an effect on dimensions with constant time evolution.