XEASY Backbone Assignment
Backbone Assignment with XEASY/UBNMR
Sequential backbone and 13CB resonance assignment is associated with mapping of SRDs identified in spin system identification onto the polypeptide sequence. This is accomplished using two (4,3)D GFT NMR experiments, that is, HNNCABCA and CABCA(CO)NHN, or using two non-GFT experiments HNNCACB/CACBCONHN.
Analysis of the (4,3)D GFT HNNCABCA and CABCA(CO)NHN spectra
HNNCABCA comprises of peaks representing both intra-residue and sequential connectivities (as in HNNCACB). These can be used to sort SRDs in sequential order, and to then assign them to specific residues in the primary structure. Since the intra-residue connectivities are often comparably weak, this experiment is routinely combined with CABCA(CO)NHN (which comprises, as CBCA(CO)NHN, sequential connectivities only).
- Peak Picking
- Go to analysis/xeasy/backbone. Edit macro getfil to import backbone spectra, sequence, prot and peaklists. Edit macro makeCabcaPeak with the sequence and prot file names. In UBNMR, run macro makeCabcaPeak to generate an extended GFT AtomList for backbone assignment which contains linear combinations of 13CA and 13CB shifts for each residue and SRD; to generate a starting peaklist for analysis of HNNCABCA and CABCA(CO)NHN by using the 15N/1HN backbone shifts of SRD-I. Intraresidue 13C shifts are assigned to SRD-I numbers, while 13C shifts of the residue preceding SRD-I are assigned to SRD-II numbers. This results in a single CABCA-peak list for the four sub-spectra of the two GFT NMR experiments. Peaks are colored according to sub-spectrum and intra- or sequential connectivity. This procedure allows one to efficiently handle sequential connectivities and ensure efficient book-keeping during the assignment process.
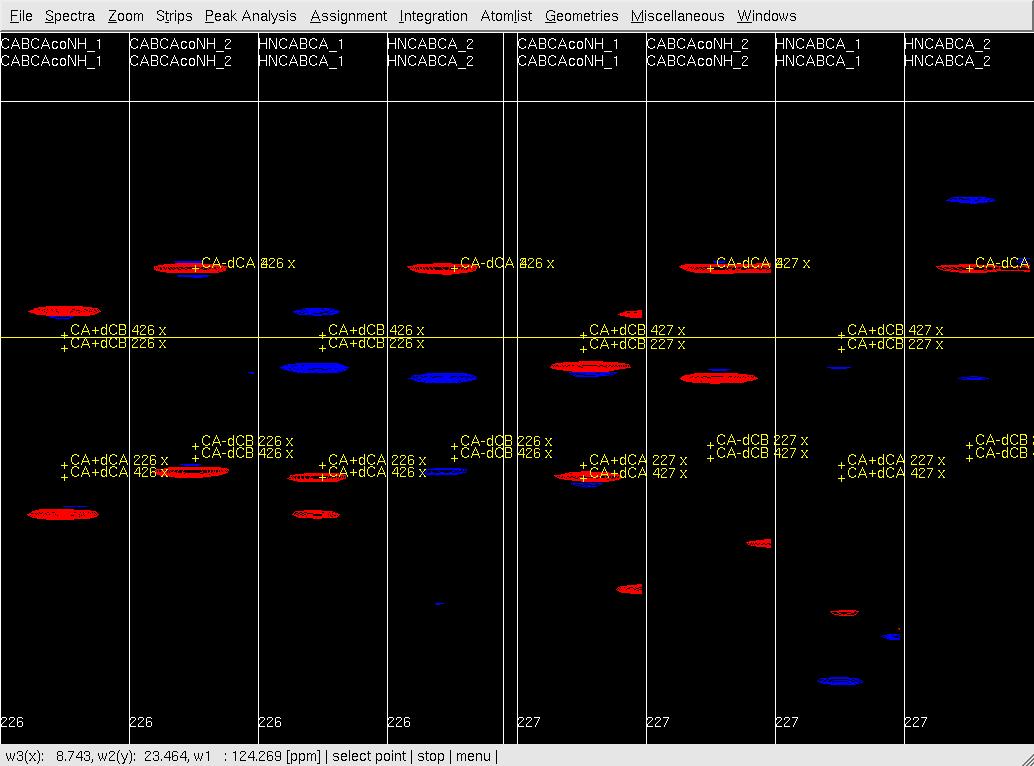
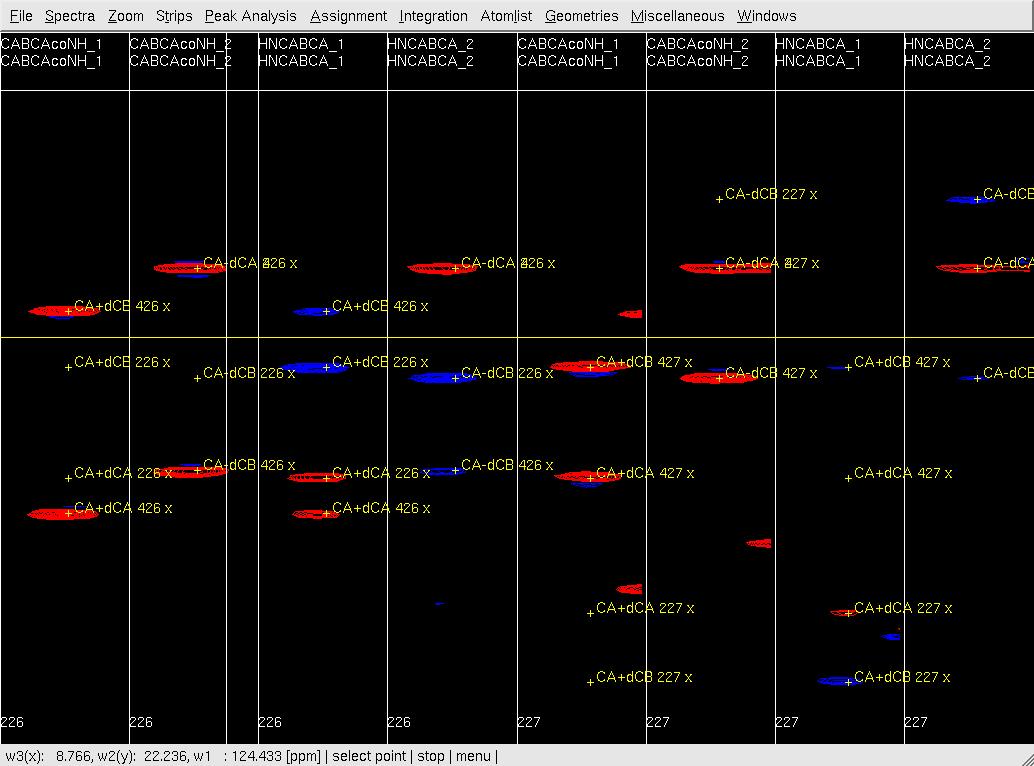
- In XEASY, use ns, ls, lc, and lp to load HNNCABCA and CABCA(CO)NHN spectra and the SequenceList, ProtonList and PeakList; use se, gs, fs and bs to sort and display strips (Figure 1A); use mr to identify and move peaks (Figure 1B); start with CABCA(CO)NHN sub-spectra and continue with HNNCABCA sub-spectra (remove unobserved peaks); use ra regularly to check on the quality of the PeakList; ac, wc and wp to save updated lists.
Figure 1: Peak picking of (4,3)D GFT HNNCABCA and CABCA(CO)NHN spectra.
A: Before peak position adjustment by mr;
B: After peak position adjustment by mr.
- Go to analysis/xeasy/backbone. Edit macro getfil to import backbone spectra, sequence, prot and peaklists. Edit macro makeCabcaPeak with the sequence and prot file names. In UBNMR, run macro makeCabcaPeak to generate an extended GFT AtomList for backbone assignment which contains linear combinations of 13CA and 13CB shifts for each residue and SRD; to generate a starting peaklist for analysis of HNNCABCA and CABCA(CO)NHN by using the 15N/1HN backbone shifts of SRD-I. Intraresidue 13C shifts are assigned to SRD-I numbers, while 13C shifts of the residue preceding SRD-I are assigned to SRD-II numbers. This results in a single CABCA-peak list for the four sub-spectra of the two GFT NMR experiments. Peaks are colored according to sub-spectrum and intra- or sequential connectivity. This procedure allows one to efficiently handle sequential connectivities and ensure efficient book-keeping during the assignment process.
- Initial Backbone Assignment from AutoAssign
- Go to /analysis/xeasy/backbone/autos, read file autos_README for instructions on how to edit the macro makeAutoList and file myprot.aat. In UBNMR run makeAutoList to generate input files for AutoAssign. Erros at this step can be corrected by looking closely at the peaks reported, moving them again if needed, and running UBNMR makeAutoList again, until all erros are eliminated. The peak ID of one simulated AutoAssign input file (myprot-hsqc.pks) correspond to the residue number of SRD-I, which is required for this protocol.
Since HNNCABCA and CABCA(CO)NHN spectra provide 4D information, it is suggested that generate the 4D peak list for AutoAssign in order to take full advantage of GFT spectra and get a better assignment results.- Click for the AutoAssign control file example myprot.aat.
- Peak pattern used in the AutoAssign input file of 3D CACB type experiments: HN(i), N(i), CA(i or i-1) or CB(i or i-1);
- Peak pattern used in the AutoAssign input file of 4D CACB type experiments: HN(i), N(i), CA(i), CA(i) or CB(i); and HN(i), N(i), CA(i-1), CA(i-1) or CB(i-1).
- Run AutoAssign several times with varying matching tolerances for CA and CB in the myprot.aat file (0.1-0.6ppm); save AutoAssign output files;
- It is suggested to use Default Execution method to run the program.
- From the main menu of AutoAssign, follow option Examine > All GSs to write output file that contains both AutoAssign assignment and the corresponding SRD-I residue numbers.
- In UBNMR, run macro AA2Xeasy to use the best or consensus AutoAssign output file to complement the SRD-I and SRD-II entries of the XEASY SequenceList with mapping numbers.
- Go to /analysis/xeasy/backbone/autos, read file autos_README for instructions on how to edit the macro makeAutoList and file myprot.aat. In UBNMR run makeAutoList to generate input files for AutoAssign. Erros at this step can be corrected by looking closely at the peaks reported, moving them again if needed, and running UBNMR makeAutoList again, until all erros are eliminated. The peak ID of one simulated AutoAssign input file (myprot-hsqc.pks) correspond to the residue number of SRD-I, which is required for this protocol.
Modified macro for AA2XEASY
#UBNMR script for converting autoassign results to xeasy format sequence init read seq ../nhsqc.seq <br> #update autoassign results to xeasy sequence update mapping aa.out 200 #200 is the number difference between self and sequencial SRD's. #write updated sequence write sequence aa.seq
- Confirming Backbone Assignment from AutoAssign in XEASY
Here two XEASY sessions are recommended for efficiency reasons, one is HNNCABCA/CABCACONHN analysis, the other is 15N NOESY analysis. For each [w1(13CA;13CAB),w3(1HN)]-strip corresponding to the 15N/1HN shifts of a given SRD-1, one expects to observe up to four [two] peaks in the two sub-spectra of (4,3)D HNNCABCA[CABCA(CO)NHN]. In the following, sequential ordering of these strips in XEASY is described. This leads to "sequential walks" in the two sub-spectra along the polypeptide chain. Sequential connectivities are confirmed in 15N-resolved NOESY Also see below the use Of 15N-resolved [1H,1H] NOESY Spectrum).
- In UBNMR, run macro makeBbNNoesy to use the 15N /1HN backbone shifts of SRD-I to generate a starting peaklist that only contains diagonal peaks for analysis of the 15N-resolved part of simultaneous 3D 15N/13Caliphatic/13Caromatic-resolved [1H,1H]-NOESY.
- In XEASY session I, ns to load the four sub-spectra of (4,3)D HNNCABCA/CABCACONHN; use ls to load the SequenceList that contains AutoAssign results; lc to load the AtomList; use lp to load the CABCA-PeakList.
- In XEASY session II, use ns to load 15N-resolved NOESY; use ls to load the SequenceList that contains AutoAssign results; use lp to load the corresponding starting peak list. There is no need to move any peaks at this time.
- In XEASY session I and II, use sn to swap fragment number and mapping; use se to sort strips; use ls and lc to load the SequenceList (with AutoAssign assignment) and the atomlist; use gs to display the strips. Now the strips are sorted in the following order: Assigned SRD residues in sequential order followed by unassigned SRD residues in SRD -1 sequential order.
- In XEASY session I and II, check and confirm sequential connectivities for the assigned SRD residues. Use ed to modify the mapping number if the assignment is wrong.
- Continue to complete the backbone assignment manually as described below
- Complete Backbone Assignment by perform sequential ordering of SRDs
Here two XEASY sessions are recommended for efficiency reasons, one is HNNCABCA/CABCACONHN analysis, the other is 15N NOESY analysis. For each [w1(13CA;13CAB),w3(1HN)]-strip corresponding to the 15N/1HN shifts of a given SRD-1, one expects to observe up to four [two] peaks in the two sub-spectra of (4,3)D HNNCABCA[CABCA(CO)NHN]. In the following, sequential ordering of these strips in XEASY is described. This leads to "sequential walks" in the two sub-spectra along the polypeptide chain. Sequential connectivities are confirmed in 15N-resolved NOESY.- In XEASY session I, use es, se, gs to select two strips exhibiting four well resolved peaks arising from a Ser, Thr, Ala residue in (4,3)D HNNCABCA or terminal residues of an assigned segment assigned from AutoAssign as a starting point; use sh to put those strips "on hold".
- In XEASY session I, use rd and pc to search for sequential neigbours.
- In XEASY session I, using fc and bc to inspect the sorted strips in order to identify the strip containing the sequential neighbor and use sh to put "on hold".
- In XEASY session II, use cd / cc to confirm sequential connectivities in 15N-resolved NOESY.
- In XEASY, repeat steps 1 to 4 until you have identified a maximal set of strips you can map to the polypeptide sequence.
- In XEASY, use ed to edit peak entries, type in "mapping numbers" which link the SRD-I and SRD-II numbers to residue numbers.
- In XEASY, repeat 5 and 6 until analysis is complete
- In XEASY, use aa, ac, ws, wc and wp to save all XEASY files before switching from SRD to sequential amino acid residue numbering using sn. This yields modified SequenceList, AtomList and PeakList which should be saved, and re-loaded and saved a second time.
- In UBNMR, run cleanBbGftProt to make a clean AtomList and SeqList by deleting extra atoms and SRDs, fixing nomenclature, and updating single-quantum 13CA and 13CA shifts.
#UBNMR macro cleanBbGftProt init read seq nhsqc.seq read prot bbgft-swapped.prot append update atom GFTatom CA update atom GFTatom CB remove SRDs write seq final-clean.seq remove GFTatoms write prot final-clean.prot
Analysis of the(4,2)D GFTHNNCABCA and CABCA(CO)NHN Spectra
In case of NMR data were collected with (4,2)D GFT HNNCABCA and CABCA(CO)NHN for backbone assignment, one can treat them the same as previously described (4,3)D GFT HNNCABCA/CABCA(CO)NHN analysis. Since the protocol of analysis of the (4,3)D GFT HNNCABCA and CABCA(CO)NHN spectra is based on analysis the stips residue by residue, it can be completely adapted to the analysis of the(4,2)D GFTHNNCABCA and CABCA(CO)NHN Spectra. Simply do the same things as described above:
- Simulating the intial peak lists;
- Adjusting peak position in strips of all residues with XEASY;
- Calculating single quantum chemical shifts from GFT shift linear combinations
- Simulating AutoAssign input files and do the initially backbone assignment by AutoAssign;
- Maually checking results from AutoAssign and completing the assignment.
Analysis of the HNNCACB/CACB(CO)NHN
In case of NMR data were collected with non-GFT NMR experiments such as HNNCACB/CACß(CO)NHN for backbone assignment, one can treat HNNCACB/CACB(CO)NHN the same as previously described (4,3)GFT HNNCABCA/CABCA(CO)NHN analysis. Simply do the same things as described in analysis of the (4,3)D GFT HNNCABCA and CABCA(CO)NHN spectra:
- Simulating the intial peak lists;
- Adjusting peak position in strips of all residues with XEASY;
- Simulating AutoAssign input files and do the initially backbone assignment by AutoAssign;
- Maually checking results from AutoAssign and completing the assignment.
Use Of 15N-resolved [1H,1H] NOESY Spectrum
Sequential NOEs observed in 15N-resolved [1H,1H] NOESY Spectrum can be used to confirm and facilitate backbone assignment. A convinient way is described here on how to do the sequential assignment by using the 15N-resolved [1H,1H] NOESY spectrum. Please check section Confirming Backbone Assignment from AutoAssign in XEASY, Complete Backbone Assignment by performing sequential ordering of SRDs and Side Chain Assignment for addtional informations.
- In UBNMR, run makeBbNNoesy to use the 15N /1HN backbone shifts of SRD-I to generate a starting peaklist that only contains diagonal peaks for analysis of the 15N-resolved part of simultaneous 3D 15N/13Caliphatic/13Caromatic-resolved [1H,1H]-NOESY.
- In XEASY, use ns to load 15N-resolved NOESY; use ls to load the SequenceList; use lp to load the corresponding starting peak list. There is no need to move any peaks at this time.
- In XEASY, use cd and cc to find sequential connectivities in 15N-resolved NOESY.
- After generating a strip list, type cd to calculated the the correlation coefficient between every two strips. he [cd command produces a pop-up window with the default values shown below.
Figure 3. The pop-up window after command cd.
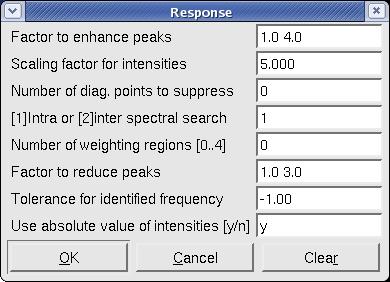
Please see the Xeasy manual for specifics regarding these parameters. The default parameters are sufficient for 15N resolved [1H, 1H]-NOESY. For searching other spectra (triple resonance, etc.) the "Factor to enhance peaks" and "Factor to reduce peaks" should both be set to "1.0 1.0". The choices for inter- and intra-spectral searches and "use absolute intensities" should also be considered. - Use gs, sf, sf or bf to display the intersted strip on the screen.
- Typing cc, then click on any position in the strip interested. Xeasy will then sort all strips according to the correlation coifficients of these strips to the clicked strips. The strips sequential residues often give the highest correlation coifficients and can be easily identified in this way. The identified sequential residues need to be confirmed from the triple resonance spectra.
- After generating a strip list, type cd to calculated the the correlation coefficient between every two strips. he [cd command produces a pop-up window with the default values shown below.
Assignment Validation
Once the assignment results were obtained from, it is required to validate the assignments. AutoAssign provides Assignment Validation Software suite (AVS) to aid the user in validating assignments. Please check the AutoAssign Tutorial at http://www-nmr.cabm.rutgers.edu/NMRsoftware/autoassign/Documentation/Help/tutorial.html for detailed usage. The four main tools in AVS are:
- missing_shifts.pl - shows which shifts in a BMRB file are missing and statistics on assignment completeness.
Usage: missing_shifts.pl [ options ] input_bmrb_file - validate_assignments.pl - performs a statistical evaluation of assigned chemical shifts in a BMRB file.
Usage: validate_assignments.pl [options] input_bmrb_file - typing_degeneracy.pl - performs a statistical evaluation on the uniqueness of the protein sequence and the uniqueness of mapped segments of spin systems.
Usage: typing_degeneracy.pl [options] - CMap Image Editor - generates graphical representations of a protein's resonance assignments and all data supporting the assignments. Please check image editor.html|Broken link for more detail.
-- Main.GaohuaLiu - 04 Feb 2007 files for download:
- makeCabcaPeak: makeCabcaPeak
- makeBbNNoesy: makeBbNNoesy
- AA2Xeasy: AA2Xeasy
- makeAutoList: makeAutoList
- myprot.aat: myprot.aat
- autos_README: autos_README